Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Cell Biology
Schlafens (SLFN) are a family of genes widely expressed in mammals, including humans and rodents. These intriguing proteins play different roles in regulating cell proliferation, cell differentiation, immune cell growth and maturation, and inhibiting viral replication. The emerging evidence is implicating Schlafens in cancer biology and chemosensitivity. Although Schlafens share common domains and a high degree of homology, different Schlafens act differently. In particular, they show specific and occasionally opposing effects in some cancer types.
- Schlafen
- cancer
- signaling
- differentiation
- invasion
- proliferation
- immune response
1. Introduction
The Schlafens (SLFN) are a novel and poorly understood family of proteins that have chiefly been investigated for their potential roles in non-malignant cell differentiation, cell proliferation, and the immune response. However, these proteins are now increasingly believed to be important in cancer. This review will briefly summarize basic Schlafen protein biology and then outline current knowledge about the role of Schlafen proteins in cancer.
Schlafens were first discovered in mice in 1998 by Schwarz et al. [1], describing the murine proteins Slfn1, Slfn2, Slfn3, and Slfn4. These proteins have subsequently been shown to be expressed in a wide range of vertebrates, including humans [2]. Mouse Slfn1 was the first discovered Schlafen and was reported to induce thymocyte cell cycle arrest (put the cell to sleep), thereby giving rise to the name of this class of proteins from the German word “Schlafen”, which means “sleep” [1]. Later, in 2000, Slfn2 was connected to Dickkopf-1 protein (DKK1) lethality in mice [3]. In 2004, Geserick et al. [4] identified and characterized another subgroup of Schlafens in mice characterized by a C-terminal sequence motif homologous to the superfamily I of DNA/RNA helicases and were identified as Slfn5, Slfn8, Slfn9, Slfn10, and Slfn14. In 2009, genomic and phylogenetic studies conducted by Bustos et al. [5] showed that Schlafens are widely expressed in mammals and identified the Schlafen genes in humans and other mammals.
Mice express ten Schlafens. These are Slfn1, Slfn1L, Slfn2, Slfn3, Slfn4, Slfn5, Slfn8, Slfn9, Slfn10 pseudogene, and Slfn14. In comparison, humans express six Schlafens. These are SLFN5, SLFN11, SLFN12, SLFN12L, SLFN13, and SLFN14 [2][6]. Schlafen genes are located on chromosome 17 in humans, while in mice they localize to chromosome 11 [2][6] (Figure 1).
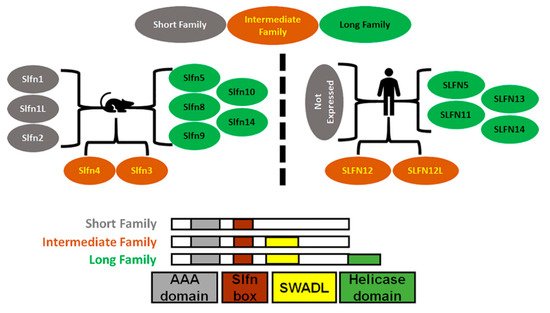
Figure 1. Diagrammatic representation of Schlafens family in mice and humans. Schlafens are classified into three families: short (grey), intermediate (orange), and long (green). Humans do not express short family Schlafens. All Schlafens share the SLFN box and putative AAA domains, while the SWADL domain is found in only intermediate and long Schlafens. Long Schlafens have an extra C-terminal helicase domain that harbors a nuclear targeting sequence.
SLFN5/Slfn5 and SLFN14/Slfn14 are the only direct orthologs between humans and mice. However, Slfn3 and Slfn4 share significant homology with SLFN12 and SLFN12L and have therefore also been identified as their orthologs [7]. Phylogenic analysis suggests Slfn8, Slfn9, and Slfn10 are orthologs to SLFN13, but there is no evidence or functional study that confirms a mouse ortholog of SLFN11.
Schlafens are expressed in diverse mammals, as well as in frogs and elephant fish. However, most research has focused on the role and function of Schlafens in mice, humans, and some viruses that express a viral ortholog of Schlafen identified as v-slfn. Schlafens were initially demonstrated to be differentially expressed in lymphoid tissue and thus believed to play a role in the maturation and activation of thymocytes [1]. However, further exploration has extended our understanding of Schlafen function to include roles in cell proliferation [8][9], cell differentiation [10][11], viral replication [12][13], cancer biology [14][15][16][17], and sensitizing cancer cells to chemotherapy [18][19][20][21][22][23].
2. Schlafens in Cancer
Although Schlafens were initially identified as proteins that influence immune cell maturation, differentiation, and responses to viral infections, recent studies have illuminated diverse potential roles for Schlafen proteins in cancer biology and malignant cell sensitivity to chemotherapy (Figure 2). The role of Schlafens in cancer has been chiefly investigated experimentally in mice and human cells and epidemiologically in humans. This review will address the effect of Schlafens in cancer in three sections. First, we will describe the role of Schlafens in the biology of different malignancies and their impact on survival, which has been chiefly investigated in humans. We will summarize the role of Schlafens in gastric cancer, malignant melanoma, lung cancer, breast cancer, gastric cancer, liver cancer, renal cancer, colorectal carcinoma, prostate cancer, central nervous system tumors, and hematological malignancies (Table 1). Second, we will summarize the role of Schlafens in the interferon immune response in cancer. Finally, we will summarize the role of Schlafens in cancer chemosensitivity to cytotoxic drugs.
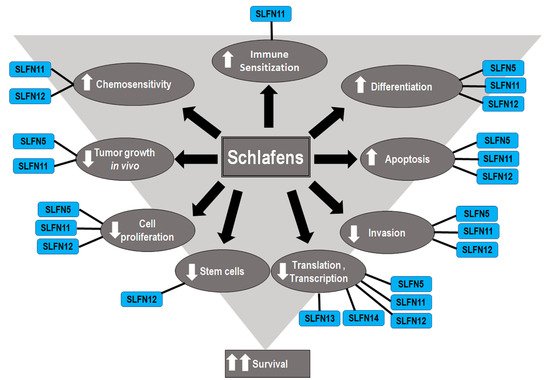
Figure 2. Diagram illustrates the identified effects of different Schlafens on cancer cell biology. The reduced cell proliferation, differentiation, invasion, and stem cells, in addition to the increased differentiation, immune sensitization, and chemosensitivity, all contribute to improved survival.
Table 1. Summary of the roles of human Schlafens in different cancer types. Each Schlafen expression level and the correlation with survival were identified. The non-cited data are derived from our analysis of publicly available tools (https://www.proteinatlas.org, accessed on 10 July 2021) and (https://kmplot.com/analysis/, accessed on 10 July 2021).
| Cancer | Schlafen | Expression Level | Survival Correlation |
|---|---|---|---|
| Malignant Melanoma | SLFN5 | Downregulated [24]. | Positive [24]. |
| SLFN11 | No significant change. | Negative. | |
| SLFN12 | No significant change. | Negative. | |
| SLFN14 | No significant change. | Positive. | |
| Renal Carcinoma | SLFN5 | Downregulated. | Positive [25]. |
| SLFN11 | Downregulated [26]. | Positive [26]. | |
| SLFN12 | Downregulated. | Negative. | |
| SLFN14 | Downregulated. | Negative. | |
| Gastric Carcinoma | SLFN5 | Upregulated [16]. | Negative [16]. |
| SLFN11 | Downregulated [27]. | Positive [27]. | |
| SLFN12 | Upregulated. | Not prognostic. | |
| SLFN14 | Downregulated. | Not prognostic. | |
| Colorectal Carcinoma | SLFN5 | No significant change. | Negative. |
| SLFN11 | Downregulated [22][26]. | Positive [22]. | |
| SLFN12 | Downregulated. | Positive. | |
| SLFN14 | Downregulated. | Not prognostic. | |
| Lung Carcinoma | SLFN5 | Downregulated [26][28]. | Positive [28]. |
| SLFN11 | Downregulated [22]. | Positive [29]. | |
| SLFN12 | Downregulated [15]. | Positive [15]. | |
| SLFN14 | Downregulated. | Positive in lung adenocarcinoma. | |
| Prostate Carcinoma | SLFN5 | Upregulated [30]. | Negative [30]. |
| SLFN11 | Upregulated in metastatic prostate cancer [31]. | No correlation to O.S., but positive correlation to rPFS [31]. | |
| SLFN12 | Downregulated. | Not prognostic. | |
| SLFN14 | Downregulated. | Not prognostic. | |
| Liver Cancer | SLFN5 | Downregulated [32]. | Positive [32]. |
| SLFN11 | Downregulated [33]. | Positive [33]. | |
| SLFN12 | Not Significant. | Negative. | |
| SLFN14 | Downregulated. | Not prognostic. | |
| Esophageal Cancer | SLFN5 | Upregulated. | No data available. |
| SLFN11 | Downregulated with age [34]. | Positive [34]. | |
| SLFN12 | Upregulated | No data available. | |
| SLFN14 | No significant change. | No data available. | |
| Breast Cancer | SLFN5 | Downregulated [35][36]. | Positive [35][36]. |
| SLFN11 | Downregulated. | Positive/negative after hormone therapy [17]. | |
| SLFN12 | Downregulated [14]. | Positive in triple negative breast cancer [14]. | |
| SLFN14 | Downregulated. | Positive. | |
| CNS Tumors | SLFN5 | Upregulated [37]. | Negative [37]. |
| SLFN11 | Upregulated [37]. | Negative [37]. | |
| SLFN12 | Upregulated [37]. | Negative [37]. | |
| SLFN13 | Upregulated [37]. | Negative [37]. | |
| Leukemia | SLFN5 | No significant change. | No data available. |
| SLFN11 | Upregulated. | No data available. | |
| SLFN12 | Upregulated. | No data available. | |
| SLFN14 | Upregulated. | No data available. |
3. Functions of Human Schlafens in Cancer
Human Schlafens act through different mechanisms to induce their effects in cancer cells. This section will briefly outline what is known about the mechanism of action of each.
SLFN5 has been reported to act through several different mechanisms. SLFN5 negatively regulates the expression of the matrix metalloproteinase (MMP) genes MMP-1 and MMP-13 [25]. SLFN5 modulates epithelial to mesenchymal transition by regulating the β-catenin signaling pathway [38][39]. SLFN5 also binds and represses STAT1 transcriptional activity, and subsequently inhibits interferon-stimulated gene response to STAT1 [37]. SLFN5 also inhibits AKT Serine/Threonine Kinase (AKT) and Glycogen Synthase Kinase 3 Beta (GSK-3β) phosphorylation [39] and upregulates Phosphatase And Tensin Homolog (PTEN) and AKT pathway by inhibiting the transcription of Zinc Finger E-Box Binding Homeobox 1 (ZEB1). Moreover, SLFN5 decreases cellular purine metabolites [35][36]. Finally, SLFN5 interacts with Activating Transcription Factor 4 (ATF4) and regulates the expression of Linker For Activation Of T Cells 1 (LAT1) (essential amino acids transporter), which in turn modulates the Mechanistic Target Of Rapamycin Kinase 1 (mTORC1) signaling pathway [30].
SLFN11 has been reported to increase chemosensitivity by five different mechanisms. First, SLFN11 is recruited to sites of DNA damage and interacts there with the Replication Protein A1 (RPA1) to inhibit checkpoint maintenance and homologous recombination repair [40]. Second, SLFN11 inhibits the translation of ataxia-telangiectasia mutated (ATM) and Rad3-related protein (ATR) (which are central components of the DNA damage response) by codon-specific translational inhibition [41]. Third, SLFN11 interacts with DDB1–CUL4CDT2 ubiquitin ligase to degrade Chromatin Licensing And DNA Replication Factor 1 (CDT1) and subsequently reactivates replication in response to chemotherapy leading to the collapse of replication forks and cell death [42]. Fourth, SLFN11 induces genome-wide chromatin accessibility at promoter regions during replication stress induced by DNA damaging agents [43]. Finally, SLFN11 is recruited to stressed replication forks, binds nascent DNA, and interacts with the replication helicase subunit (MCM3) without inhibiting initiation or loading of replication factor Cell Division Cycle 45 (CDC45) or the proliferating cell nuclear antigen (PCNA). This then allows SLFN11 to subsequently block replication in response to replication stress independently from ATR within 4 hours of induction of DNA damage [44].
SLFN12 binds Serpin B12 and subsequently modulates the deubiquitylases USP14 and UCHL5 to inhibit the proteasomal degradation of differentiation transcription factors such as cdx2 [45]. SLFN12 inhibits the translation of the oncogenes ZEB1 [14] and C-Myc [15]. SLFN12 is stabilized by interaction with Phosphodiesterase 3A (PDE3A) and is recruited and binds the ribosomes to exclude signal recognition peptides and subsequently inhibits translation [46]. In addition to these effects, SLFN12 has recently been identified as an RNAse that digests ribosomal RNA [47].
SLFN13, in contrast to other long Schlafens, is localized to the cytoplasm and acts as tRNA/rRNA-endoribonuclease that digests both tRNAs and rRNAs with a preference for tRNA, which inhibits protein synthesis [48]. In contrast, the long SLFN14 is a ribosome-associated nuclear protein that binds to the ribosomal subunits and cleaves RNA, in particular rRNA and ribosome-associated mRNA, to control mRNA turnover and protein translation [49].
4. Conclusions
Although it has been more than twenty years since Schlafens were discovered, they remain intriguing proteins and are still enigmas. The exploration of the role of Schlafens in cancers is still in its early stages and evidence predicts a considerable role for Schlafens in tumorigenesis, as biomarkers and predictors for chemotherapy, and as possible targets for drugs.
The role of Schlafens is not universal, and each Schlafen has a distinct specific mechanism through which it affects cancer cell biology. Interestingly, this mechanism is not alike in all cancer types/subtypes. Instead, Schlafens demonstrate different effects in different subtypes of tumors of the same organ.
The complex signaling networks through which Schlafens are reported to act in cancer urges extensive future characterization and exploration of each Schlafen in the different cancer subtypes, which will eventually push us closer to better understand cancers in humans.
This entry is adapted from the peer-reviewed paper 10.3390/cells10092238
References
- Schwarz, D.A.; Katayama, C.D.; Hedrick, S.M. Schlafen, a New Family of Growth Regulatory Genes that Affect Thymocyte Development. Immunity 1998, 9, 657–668.
- Chen, J.; Kuhn, L.A. Deciphering the three-domain architecture in schlafens and the structures and roles of human schlafen12 and serpinB12 in transcriptional regulation. J. Mol. Graph. Model. 2019, 90, 59–76.
- Cohen-Tannoudji, M.; Vandormael-Pournin, S.; Le Bras, S.; Coumailleau, F.; Babinet, C.; Baldacci, P. A 2-Mb YAC/BAC-Based Physical Map of the Ovum Mutant (Om) Locus Region on Mouse Chromosome 11. Genomics 2000, 68, 273–282.
- Geserick, P.; Kaiser, F.; Klemm, U.; Kaufmann, S.H.E.; Zerrahn, J. Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int. Immunol. 2004, 16, 1535–1548.
- Bustos, O.; Naik, S.; Ayers, G.; Casola, C.; Perez-Lamigueiro, M.A.; Chippindale, P.T.; Pritham, E.J.; de la Casa-Esperón, E. Evolution of the Schlafen genes, a gene family associated with embryonic lethality, meiotic drive, immune processes and orthopoxvirus virulence. Gene 2009, 447, 1–11.
- Liu, F.; Zhou, P.; Wang, Q.; Zhang, M.; Li, D. The Schlafen family: Complex roles in different cell types and virus replication. Cell Biol. Int. 2018, 42, 2–8.
- De La Casa-Esperón, E. From mammals to viruses: The Schlafen genes in developmental, proliferative and immune processes. Biomol. Concepts 2011, 2, 159–169.
- Brady, G.; Boggan, L.; Bowie, A.; O’Neill, L. Schlafen-1 Causes a Cell Cycle Arrest by Inhibiting Induction of Cyclin D1. J. Biol. Chem. 2005, 280, 30723–30734.
- Kuang, C.-Y.; Yang, T.-H.; Zhang, Y.; Zhang, L.; Wu, Q. Schlafen 1 Inhibits the Proliferation and Tube Formation of Endothelial Progenitor Cells. PLoS ONE 2014, 9, e109711.
- Basson, M.D.; Wang, Q.; Chaturvedi, L.S.; Vomhof-DeKrey, E. Schlafen 12 Promotes Human Intestinal Epithelial Differentiation via Serpin B12 Modulation of the Deubiquitylation of Transcription Factors Such as CDX2. Gastroenterology 2018, 154, S180.
- Oh, P.-S.; Patel, V.B.; Sanders, M.A.; Kanwar, S.; Yu, Y.; Nautiyal, J.; Patel, B.B.; Majumdar, A.P.N. Schlafen-3 decreases cancer stem cell marker expression and autocrine/juxtacrine signaling in FOLFOX-resistant colon cancer cells. Am. J. Physiol. Liver Physiol. 2011, 301, G347–G355.
- Abdel-Mohsen, M.; Raposo, R.A.S.; Deng, X.; Li, M.; Liegler, T.; Sinclair, E.; Salama, M.S.; Ghanem, A.H.E.-D.; Hoh, R.; Wong, J.K.; et al. Expression profile of host restriction factors in HIV-1 elite controllers. Retrovirology 2013, 10, 106.
- Abdel-Mohsen, M.; Wang, C.; Strain, M.C.; Lada, S.M.; Deng, X.; Cockerham, L.R.; Pilcher, C.D.; Hecht, F.; Liegler, T.; Richman, D.D.; et al. Select host restriction factors are associated with HIV persistence during antiretroviral therapy. AIDS 2015, 29, 411–420.
- Al-Marsoummi, S.; Vomhof-DeKrey, E.; Basson, M.D. Schlafen12 Reduces the Aggressiveness of Triple Negative Breast Cancer through Post-Transcriptional Regulation of ZEB1 That Drives Stem Cell Differentiation. Cell. Physiol. Biochem. 2019, 53, 999–1014.
- Al-Marsoummi, S.; Pacella, J.; Dockter, K.; Soderberg, M.; Singhal, S.K.; Vomhof-DeKrey, E.E.; Basson, M.D. Schlafen 12 Is Prognostically Favorable and Reduces C-Myc and Proliferation in Lung Adenocarcinoma but Not in Lung Squamous Cell Carcinoma. Cancers 2020, 12, 2738.
- Nápoles, O.C.; Tsao, A.C.; Sanz-Anquela, J.M.; Sala, N.; Bonet, C.; Pardo, M.L.; Ding, L.; Simo, O.; Saqui-Salces, M.; Blanco, V.P.; et al. SCHLAFEN 5 expression correlates with intestinal metaplasia that progresses to gastric cancer. J. Gastroenterol. 2017, 52, 39–49.
- Isnaldi, E.; Ferraioli, D.; Ferrando, L.; Brohée, S.; Ferrando, F.; Fregatti, P.; Bedognetti, D.; Ballestrero, A.; Zoppoli, G. Schlafen-11 expression is associated with immune signatures and basal-like phenotype in breast cancer. Breast Cancer Res. Treat. 2019, 177, 335–343.
- Tian, L.; Song, S.; Liu, X.; Wang, Y.; Xu, X.; Hu, Y.; Xu, J. Schlafen-11 sensitizes colorectal carcinoma cells to irinotecan. Anti-Cancer Drugs 2014, 25, 1175–1181.
- Zoppoli, G.; Regairaz, M.; Leo, E.; Reinhold, W.C.; Varma, S.; Ballestrero, A.; Doroshow, J.H.; Pommier, Y. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc. Natl. Acad. Sci. USA 2012, 109, 15030–15035.
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607.
- Nogales, V.; Reinhold, W.; Varma, S.; Cardus, A.M.; Moutinho, C.; Moran, S.; Heyn, H.; Sebio, A.; Barnadas, A.; Pommier, Y.; et al. Epigenetic inactivation of the putative DNA/RNA helicase SLFN11 in human cancer confers resistance to platinum drugs. Oncotarget 2016, 7, 3084–3097.
- He, T.; Zhang, M.; Zheng, R.; Zheng, S.; Linghu, E.; Herman, J.G.; Guo, M. Methylation ofSLFN11is a marker of poor prognosis and cisplatin resistance in colorectal cancer. Epigenomics 2017, 9, 849–862.
- Stewart, C.A.; Tong, P.; Cardnell, R.J.; Sen, T.; Li, L.; Gay, C.M.; Masrorpour, F.; Fan, Y.; Bara, R.O.; Feng, Y.; et al. Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget 2017, 8, 28575–28587.
- Katsoulidis, E.; Mavrommatis, E.; Woodard, J.; Shields, M.A.; Sassano, A.; Carayol, N.; Sawicki, K.T.; Munshi, H.G.; Platanias, L.C. Role of interferon α (IFNα)-inducible Schlafen-5 in regulation of anchorage-independent growth and invasion of malignant melanoma cells. J. Biol. Chem. 2010, 285, 40333–40341.
- Sassano, A.; Mavrommatis, E.; Arslan, A.D.; Kroczynska, B.; Beauchamp, E.M.; Khuon, S.; Chew, T.-L.; Green, K.J.; Munshi, H.G.; Verma, A.K.; et al. Human Schlafen 5 (SLFN5) Is a Regulator of Motility and Invasiveness of Renal Cell Carcinoma Cells. Mol. Cell. Biol. 2015, 35, 2684–2698.
- Takashima, T.; Sakamoto, N.; Murai, J.; Taniyama, D.; Honma, R.; Ukai, S.; Maruyama, R.; Kuraoka, K.; Rajapakse, V.N.; Pommier, Y.; et al. Immunohistochemical analysis of SLFN11 expression uncovers potential non-responders to DNA-damaging agents overlooked by tissue RNA-seq. Virchows Arch. 2021, 478, 569–579.
- Takashima, T.; Taniyama, D.; Sakamoto, N.; Yasumoto, M.; Asai, R.; Hattori, T.; Honma, R.; Thang, P.Q.; Ukai, S.; Maruyama, R.; et al. Schlafen 11 predicts response to platinum-based chemotherapy in gastric cancers. Br. J. Cancer 2021.
- Yang, X.; Peng, R.; Peng, H.; Zhang, Q.; Long, G.; Wu, G.; Liang, Z. The association between SLFN5 expression and the prognosis of non-small cell lung cancer. Int. J. Clin. Exp. Med. 2019, 12, 682–689.
- Shee, K.; Wells, J.D.; Jiang, A.; Miller, T.W. Integrated pan-cancer gene expression and drug sensitivity analysis reveals SLFN11 mRNA as a solid tumor biomarker predictive of sensitivity to DNA-damaging chemotherapy. PLoS One 2019, 14, e0224267.
- Martinez, R.S.; Salji, M.J.; Rushworth, L.; Ntala, C.; Rodriguez Blanco, G.; Hedley, A.; Clark, W.; Peixoto, P.; Hervouet, E.; Renaude, E.; et al. SLFN5 regulates LAT1-mediated mTOR activation in castration-resistant prostate cancer. Cancer Res. 2021.
- Conteduca, V.; Ku, S.-Y.; Puca, L.; Slade, M.; Fernandez, L.; Hess, J.; Bareja, R.; Vlachostergios, P.J.; Sigouros, M.; Mosquera, J.M.; et al. SLFN11 Expression in Advanced Prostate Cancer and Response to Platinum-based Chemotherapy. Mol. Cancer Ther. 2020, 19, 1157–1164.
- Peng, H.; Yang, M.; Xu, X.M.; She, S.; Ran, X.P.; Li, S.Y.; Hu, H.D.; Ren, H.; Hu, P. Decreased expression of schlafen5 (SLFN5) correlates with unfavorable survival in human hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2016, 9, 7014–7096.
- Zhou, C.; Liu, C.; Liu, W.; Chen, W.; Yin, Y.; Li, C.-W.; Hsu, J.L.; Sun, J.; Zhou, Q.; Li, H.; et al. SLFN11 inhibits hepatocellular carcinoma tumorigenesis and metastasis by targeting RPS4X via mTOR pathway. Theranostics 2020, 10, 4627–4643.
- Kagami, T.; Yamade, M.; Suzuki, T.; Uotani, T.; Tani, S.; Hamaya, Y.; Iwaizumi, M.; Osawa, S.; Sugimoto, K.; Miyajima, H.; et al. The first evidence for SLFN11 expression as an independent prognostic factor for patients with esophageal cancer after chemoradiotherapy. BMC Cancer 2020, 20, 1123.
- Wan, G.; Zhu, J.; Gu, X.; Yang, Y.; Liu, Y.; Wang, Z.; Zhao, Y.; Wu, H.; Huang, G.; Lu, C. Human Schlafen 5 regulates reversible epithelial and mesenchymal transitions in breast cancer by suppression of ZEB1 transcription. Br. J. Cancer 2020, 123, 633–643.
- Gu, X.; Wan, G.; Yang, Y.; Liu, Y.; Yang, X.; Zheng, Y.; Jiang, L. SLFN5 influences proliferation and apoptosis by upregulating PTEN transcription via ZEB1 and inhibits the purine metabolic pathway in breast cancer. Am. J. Cancer Res. 2020, 10, 2832–2850.
- Arslan, A.D.; Sassano, A.; Saleiro, D.; Lisowski, P.; Kosciuczuk, E.M.; Fischietti, M.; Eckerdt, F.; Fish, E.N.; Platanias, L.C. Human SLFN5 is a transcriptional co-repressor of STAT1-mediated interferon responses and promotes the malignant phenotype in glioblastoma. Oncogene 2017, 36, 6006–6019.
- Guo, L.; Liu, Z.; Tang, X. Overexpression of SLFN5 induced the epithelial-mesenchymal transition in human lung cancer cell line A549 through β-catenin/Snail/E-cadherin pathway. Eur. J. Pharmacol. 2019, 862, 172630.
- Wan, G.; Liu, Y.; Zhu, J.; Guo, L.; Li, C.; Yang, Y.; Gu, X.; Deng, L.-L.; Lu, C. SLFN5 suppresses cancer cell migration and invasion by inhibiting MT1-MMP expression via AKT/GSK-3β/β-catenin pathway. Cell. Signal. 2019, 59, 1–12.
- Mu, Y.; Lou, J.; Srivastava, M.; Zhao, B.; Feng, X.; Liu, T.; Chen, J.; Huang, J. SLFN11 inhibits checkpoint maintenance and homologous recombination repair. EMBO Rep. 2016, 17, 94–109.
- Li, M.; Kao, E.; Malone, D.; Gao, X.; Wang, J.Y.J.; David, M. DNA damage-induced cell death relies on SLFN11-dependent cleavage of distinct type II tRNAs. Nat. Struct. Mol. Biol. 2018, 25, 1047–1058.
- Jo, U.; Murai, Y.; Chakka, S.; Chen, L.; Cheng, K.; Murai, J.; Saha, L.K.; Miller Jenkins, L.M.; Pommier, Y. SLFN11 promotes CDT1 degradation by CUL4 in response to replicative DNA damage, while its absence leads to synthetic lethality with ATR/CHK1 inhibitors. Proc. Natl. Acad. Sci. USA 2021, 118, e2015654118.
- Murai, J.; Zhang, H.; Pongor, L.; Tang, S.W.; Jo, U.; Moribe, F.; Ma, Y.; Tomita, M.; Pommier, Y. Chromatin Remodeling and Immediate Early Gene Activation by SLFN11 in Response to Replication Stress. Cell Rep. 2020, 30, 4137–4151.
- Murai, J.; Tang, S.W.; Leo, E.; Baechler, S.A.; Redon, C.E.; Zhang, H.; Al Abo, M.; Rajapakse, V.N.; Nakamura, E.; Jenkins, L.M.M.; et al. SLFN11 Blocks Stressed Replication Forks Independently of ATR. Mol. Cell 2018, 69, 371–384.
- Basson, M.D.; Wang, Q.; Chaturvedi, L.S.; More, S.; Vomhof-DeKrey, E.E.; Al-Marsoummi, S.; Sun, K.; Kuhn, L.A.; Kovalenko, P.; Kiupel, M. Schlafen 12 Interaction with Serpin B12 and Deubiquitylases Drives Human Enterocyte Differentiation. Cell. Physiol. Biochem. 2018, 48, 1274–1290.
- Li, D.; Chen, J.; Ai, Y.; Gu, X.; Li, L.; Che, D.; Jiang, Z.; Li, L.; Chen, S.; Huang, H.; et al. Estrogen-Related Hormones Induce Apoptosis by Stabilizing Schlafen-12 Protein Turnover. Mol. Cell 2019, 75, 1103–1116.
- Garvie, C.W.; Wu, X.; Papanastasiou, M.; Lee, S.; Fuller, J.; Schnitzler, G.R.; Horner, S.W.; Baker, A.; Zhang, T.; Mullahoo, J.P.; et al. Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase. Nat. Commun. 2021, 12, 4375.
- Yang, J.-Y.; Deng, X.-Y.; Li, Y.-S.; Ma, X.; Feng, J.-X.; Yu, B.; Chen, Y.; Luo, Y.-L.; Wang, X.; Chen, M.-L.; et al. Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control. Nat. Commun. 2018, 9, 1–13.
- Pisareva, V.P.; Muslimov, I.A.; Tcherepanov, A.; Pisarev, A.V. Characterization of Novel Ribosome-Associated Endoribonuclease SLFN14 from Rabbit Reticulocytes. Biochemistry 2015, 54, 3286–3301.
This entry is offline, you can click here to edit this entry!
