The SARS-CoV-2 main protease (Mpro) is one of the molecular targets for drug design. Effective vaccines have been identified as a long-term solution but the rate at which they are being administered is slow in several countries, and mutations of SARS-CoV-2 could render them less effective.
1. Introduction
Viral infection is one of the major challenges faced by human health, and many viral diseases are correlated with high morbidity and mortality rates in humans. Previously, viral diseases such as influenza, dengue, HIV, and coronaviruses have resulted in epidemics or global pandemics, claiming many lives. Currently, the world is battling with coronavirus diseases 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) which has resulted in approximately 4 million deaths as of July 2021. The emergence of variants of this disease has also been challenging to the developed vaccines, and, as it stands, there are no effective therapeutic interventions against this disease. Proteins associated with viral infection can serve as molecular targets for disease prevention and treatment. Molecules involved in viral DNA replication and protein synthesis can be pivotal to these processes. Targets such as Papain-like protease (PLpro), 3-chymotrypsin like protease (3CLPro) also called main protease (M
pro), RNA-dependent RNA polymerase (RdRp), and helicase have been reported as potential points of treatment development for SARS-CoV-2 infection [
1,
2,
3]. The SARS-CoV-2 spike protein and angiotensin-converting enzyme-2 (ACE-2) were also identified as promising targets for disease prevention [
4,
5,
6]. The non-structural proteins essential for the replication of viral particles are specifically generated by PLpro and M
pro, defining their roles in viral replication and outlining their inhibition as potential anti-SARS-CoV-2 treatments [
7,
8,
9].
Biologically active molecules have been screened against M
pro as either repurposed drugs or in the process of lead identification. Muhammad et al. [
10] screened a library of phytochemicals against M
pro and revealed the potential usage of molecules from natural sources as anti-COVID-19 druggable candidates. In another study, bioactive medicinal plants were assayed against M
pro using in silico docking and pharmacological screening. Selected molecules from alkaloids and terpenoids were identified as inhibitors of this target, with a highly conserved inhibitory pattern to both SARS-CoV-2 and SARS-CoV [
11]. Moreover, nucleopeptides and Opuntia-derived phytochemicals were suggested as M
pro potential inhibitors [
12,
13]. Therefore, the inhibition of protein activity of M
pro may potentially suppress coronavirus transmission.
Natural products of low molecular weight from plant sources are potent therapeutic agents for many diseases and some of these agents possess antiviral properties. While many natural products are fundamentally utilized as crude extracts, the purification of their active ingredients is essential for the prediction of their properties associated with the pharmacokinetics and pharmacodynamics of a drug molecule.
Accumulating evidence suggests that plant chemicals, for example, polyphenols and their functional derivatives such as flavonoids, saponins, and lignans can alter cellular functions, membrane permeability, and viral replication [
14]. The role of phytochemicals has also been implicated in cell migration and proliferation, metabolism regulation (phytosterol, flavanols, anthocyanidins, cinnamic acids, etc.) [
15,
16], inflammatory processes (quercetin, kaempferol, etc.) [
17], redox modulation (phenolics, curcumin, resveratrol, etc.) [
18,
19], and angiogenesis (astaxanthin) [
20]. Ethanolic extract of
Ficus benjamina has been shown to contain some active compounds such as rutin, kaempferol 3-O-rutinoside, and kaempferol 3-O-robinobioside, which were effective against herpes simplex [
21]. Equally, the phytochemicals homolycorine and 2-O-acetyllycorine isolated from
Leucojum vernum were shown to be effective against HIV-1 [
22]. Rutin is a glycosylated flavonoid with a 3-rutinoside substitution. Its antiviral activity has been studied against avian influenza virus [
23], herpes simplex [
21], and parainfluenza-3 virus [
24].
The world population is largely dependent on therapies from plant origin [
25]. Compounds isolated from this source have little or no side effects with high biological specificity, chemical diversity, and targets multiple host sites by diverse pathways with negligible cost [
26]. It was previously demonstrated that flavonoids, among other active compounds from plant sources, have been used for the treatment of HIV, herpes simplex, and influenza viruses, due to their antiviral properties [
27]. These compounds inhibit viral replication, proteases, and reverse transcription [
28]. Compounds such as quercetin, rutin, and myricetin have also been identified with similar properties [
29].
2. ADME/Tox Prediction
The pharmacokinetics and toxicological properties of the ligands (
Table 1 and
Table 2) were analyzed according to previous methods to investigate how molecules can access the target site of M
pro after entering the bloodstream. This analysis is also crucial for analyzing the efficacy of molecules [
42,
43]. All parameters were within the ROF cut-off range for the test compounds and present no bystander toxicity effects since toxicity is the main task in developing new medications. Ames toxicity, carcinogenic properties, and rat acute toxicity were predicted in the current investigation.
Table 1. Properties of the selected compounds.
| Compound |
ID a |
M.W b |
ROF c |
QplogHERG d |
QplogPoW e |
QplogKP f |
Donor HB |
Acceptor HB |
QplogS g |
QplogBB h |
| Quercetin 3-O-Neohesperidoside |
5748416 |
610.5 |
2 |
−6.449 |
−1.998 |
−6.423 |
9 |
20.55 |
−2.932 |
−4.728 |
| Myricetin 3-Rutinoside |
44259428 |
626.5 |
2 |
−6.394 |
−2.455 |
−5.583 |
10 |
21.3 |
−2.341 |
−4.306 |
| Quercetin 3-Rhamnoside |
5353915 |
448.3 |
2 |
−5.451 |
−0.55 |
−6.101 |
6 |
12.05 |
−3.196 |
−3.312 |
| Rutin |
5280805 |
610.5 |
2 |
−5.238 |
−2.495 |
−7.251 |
9 |
20.55 |
−2.175 |
−4.503 |
| Myricitrin |
5281673 |
464.3 |
2 |
−5.463 |
−1.045 |
−6.589 |
7 |
12.8 |
−2.779 |
−3.48 |
a Compound ID from PubChem database. b Formula weight of the compounds (acceptable range: 130.0–725 g/mol). c Number of permissible violations of Lipinski’s rule of five (acceptable range: maximum is 4). d Predicted IC50 value for blockage of HERG K+ channels (concern below −5.0). e Predicted octanol/water partition coefficient log p (acceptable range: −2.0 to 6.5). f Predicted skin permeability, log Kp (acceptable range: −8.0 to −1.0). g Predicted aqueous solubility; S in mol/L (acceptable range: −6.5 to 0.5). h Predicted brain/blood partition coefficient (acceptable range: −3.0 to 1.2). Donor HB (≤10); Acceptor HB (≤5).
Table 2. Toxicity analysis of the selected compounds predicted by AdmetSAR.
| Compound |
ID |
Ames Toxicity |
Carcinogens |
Acute Oral Toxicity |
Rat Acute Toxicity |
| Quercetin 3-O-Neohesperidoside |
5748416 |
AT |
NC |
III |
2.2619 |
| Myricetin 3-Rutinoside |
44259428 |
NAT |
NC |
III |
2.4984 |
| Quercetin 3-Rhamnoside |
5353915 |
NAT |
NC |
III |
2.5458 |
| Rutin |
5280805 |
NAT |
NC |
III |
2.4984 |
| Myricitrin |
5281673 |
NAT |
NC |
III |
2.5458 |
Note: AT: Ames toxic NAT: Non Ames toxic; NC: Non-carcinogenic; Category-III means (500 mg/kg > LD50 < 5000 mg/kg).
3. Docking Calculations
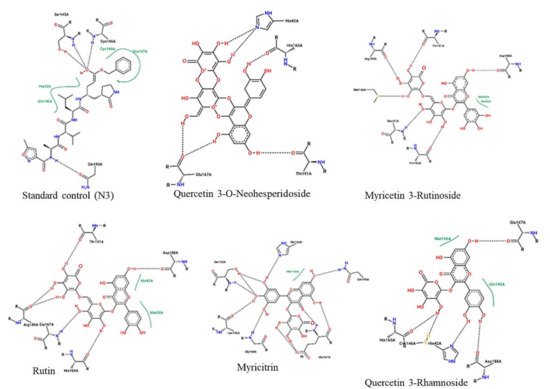
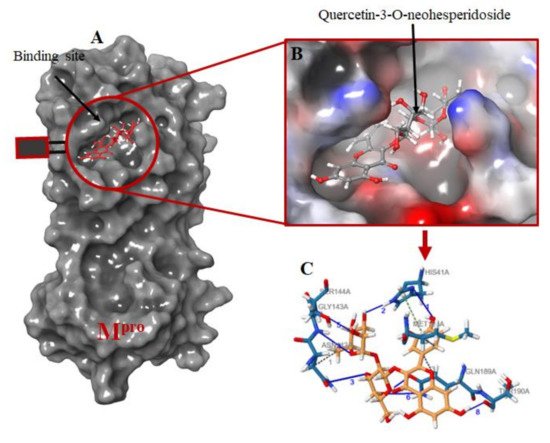
The protein-ligand interactions for all the complexes after the docking procedure were produced by Proteins Plus at
https://proteins.plus/ (accessed on 1 August 2021) as depicted in
Figure 1. The model representation of the best pose against decoy poses was also presented using M
pro-Quercetin-3-O-Neohesperidoside (
Figure 2). The binding properties, such as the scoring functions of Autodock Vina and MM/GBSA, the number of hydrogen bond integrations, and types of residues and their distances (Å) are tabulated in
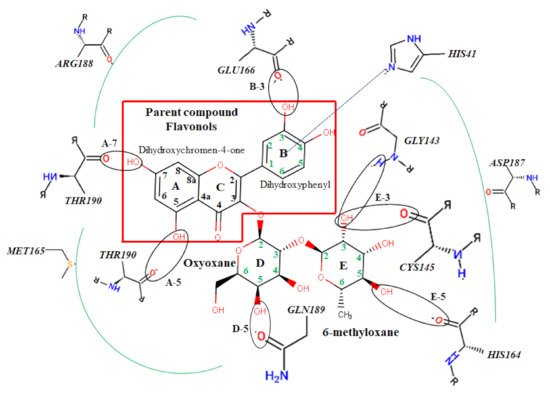
Table 3. In addition, the possible residue interaction crucial to the inhibition process of M
pro by these flavonoids was proposed in
Figure 3.
Figure 1. 2-dimentional protein–ligand interactions created for docked ligands and N3 into the active site of Mpro ranked according to their binding energies. Black bond interactions showed H-bonds between the atoms of the ligands and the residues of the receptor.
Figure 2. Molecular docking of the ligands into Mpro binding site. (A) the docking of the selected ligands in Mpro binding pocket; (B) Quercetin-3-O-Neohesperidoside seats perfectly in the Mpro binding pocket; (C) interacting atoms of Quercetin-3-O-Neohesperidoside and Mpro residues in the binding pocket. Note: blue lines indicate hydrogen bond interaction; green dotted lines indicate pi-stacking, while gray dotted lines depict hydrophobic interactions.
Figure 3. Atomic interaction of flavonoids with Mpro residues. While several residues form hydrophobic interactions with the ligands, other interactions such as hydrogen, π-cation, and π-π stacking were also involved in the inhibition mechanism of Mpro.
Table 3. Binding energies of flavonoids docked against Mpro.
| Name |
Dock Score |
∆G Bind |
H-Bond |
Residues (Å) |
Other Bond (Å) |
| Standard |
−16.5 |
−80.88 |
6 |
CYS145 (2.13), LEU141 (2.76), PHE140 (2.02), GLU166 (1.68,1.83, 2.05) |
Salt bridges (2) |
| Quercetin 3-O-Neohesperidoside |
−16.8 |
−87.60 |
5 |
GLY143 (2.76), CYS145 (2.11), GLN189 (2.11), THR190 (1.76), HIS41 (2.30) |
π-π stacking HIS41 (1.49) |
| Myricetin 3-Rutinoside |
−12.9 |
−87.50 |
7 |
CYS145 (2.08), ASN142 (1.75), GLU166 (1.98), THR190 (2.21), ARG188 (1.97), HIS164 (1.90,1.98) |
|
| Quercetin 3-Rhamnoside |
−10.3 |
−80.17 |
4 |
LEU141 (1.49), THR190 (1.78), GLU166 (2.01), HIS164 (1.81) |
|
| Rutin |
−10.0 |
−58.95 |
6 |
THR190 (1.83), HIS41 (2.08), GLY143 (2.39,1.89), ASN142 (1.92), LEU141 (2.10) |
- |
| Myricitrin |
−9.1 |
−49.22 |
3 |
CYS145 (2.51), ASN142 (2.04), THR190 (1.83) |
π-π stacking HIS41 (5.37) |
4. Molecular Dynamic Simulation
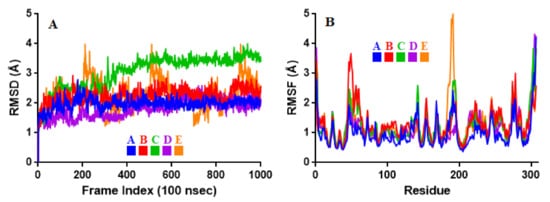
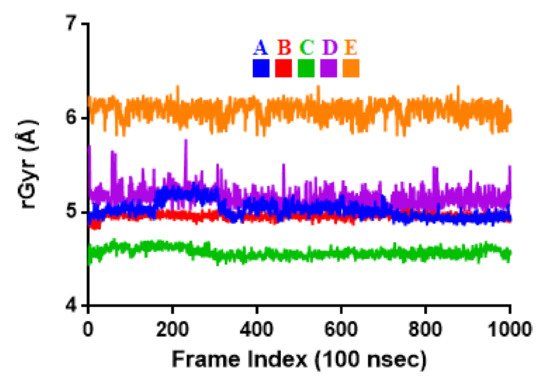
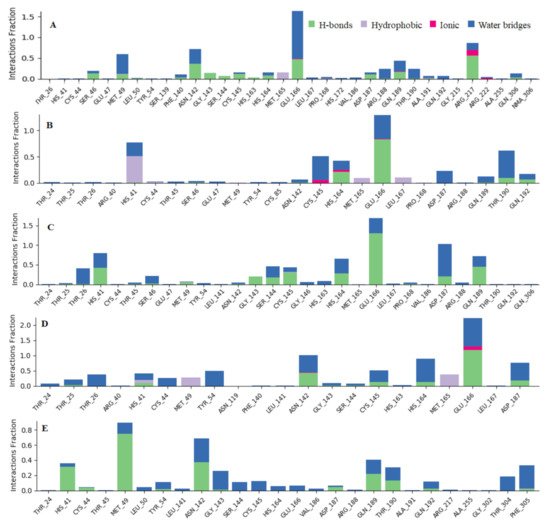
To associate structural and mechanistic information with experimental data, the MDs were carried out. The Mpro-ligand complexes were computationally simulated for 100 ns to decipher the complex stability and dynamic behavior as presented in Figure 4, Figure 5 and Figure 6, and Table 2. The root-mean-square deviation (RMSD) of the five ligands were plotted against 1000 frame indexes for 100 ns (Figure 4a). The root-mean-square fluctuation (RMSF) of the C-alpha of the protein complexed with these ligands was also plotted against residues (Figure 4b). The ligand properties were also taken into account by plotting the radius of gyration (rGyr) for all five complexes against the frame index over the 100 ns simulation time (Figure 5). The simulation properties were calculated as the mean ± SD for RMSD, RMSF, and rGyr (Table 4). Finally, protein interactions with the ligands were monitored throughout the simulation. These interactions were categorized by type and summarized as shown in Figure 6.
Figure 4. Stability and flexibility of the selected flavonoid derivatives complexed with Mpro over the course of 100 ns. (A) is the RMS deviation and (B) is the RMS functions of the Cα of each complexes. Color codes denote: Quercetin-3-O-Neohesperidoside (A), Quercetin 3-Rhamnoside (B), Myricitrin (C), Rutin (D), and Myricetin 3-Rutinoside (E).
Figure 5. Radius of gyration (rGyr) plot for Mpro and the selected flavonoid derivatives within the simulation time of 100 ns. Color codes denote: Quercetin-3-O-Neohesperidoside (A), Quercetin 3-Rhamnoside (B), Myricitrin (C), Rutin (D), and Myricetin 3-Rutinoside (E).
Figure 6. Observed Mpro-ligands interaction during the 100 ns MD simulation. Interactions include; hydrogen bonds, hydrophobic, ionic and water bridges. Letter codes indicate: Quercetin-3-O-Neohesperidoside (A), Quercetin 3-Rhamnoside (B), Myricitrin (C), Rutin (D), and Myricetin 3-Rutinoside (E).
Table 4. The simulation properties of the complexes.
| Properties |
A |
B |
C |
D |
E |
| RMSD |
1.98 ± 0.19 |
2.25 ± 0.26 |
3.05 ± 0.57 |
1.81 ± 0.30 |
2.26 ± 0.51 |
| RMSF |
1.00 ± 0.51 |
1.31 ± 0.55 |
1.27 ± 0.58 |
1.15 ± 0.58 |
1.25 ± 0.65 |
| rGyr |
5.03 ± 0.09 |
4.96 ± 0.03 |
4.57 ± 0.05 |
5.15 ± 0.10 |
6.09 ± 0.09 |
Values represent the mean ± SD of 1000 frame index replicates. Letter codes: Quercetin-3-O-Neohesperidoside (A), Quercetin 3-Rhamnoside (B), Myricitrin (C), Rutin (D), and Myricetin 3-Rutinoside (E). Values are in Å.
This entry is adapted from the peer-reviewed paper 10.3390/ijms22179431