During liver organogenesis, cellular transcriptional profiles are constantly reshaped by the action of hepatic transcriptional regulators, including FoxA1-3, GATA4/6, HNF1α/β, HNF4α, HNF6, OC-2, C/EBPα/β, Hex, and Prox1. These factors are crucial for the activation of hepatic genes. The initial opening of highly condensed chromatin is executed by a special class of transcription factors known as pioneer factors. This is followed by the progressive recruitment of chromatin modifiers and the stable or transient binding of other transcription factors, which lead to the gradual deposition of activating histone modifications and the broadening of active chromatin domains. The resulting permissive chromatin state facilitates the assembly of the pre-initiation complex (PIC) and promotes transcriptional initiation.
- liver
- transcription factor
- chromatin
- development
- bookmarking
- gene expression
1. Introduction
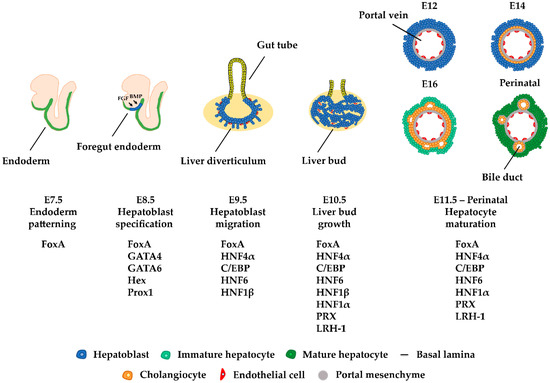
The liver participates in a variety of crucial biological processes such as hemopoiesis during embryonic life and metabolism, glycogen storage, detoxification, plasma protein secretion, acute phase reaction, and hormonal homeostasis in adulthood. The major cell type of the liver is the hepatocyte, which arises from endodermal precursors through a complex multistep differentiation process. During hepatocyte differentiation, the gene expression pattern of each intermediate cell type is generated by the action of transcription factors, which bind to the regulatory regions of their target genes and activate transcription at specific times during development. Developmental cell fate decisions are determined by cell-to-cell communication and the action of complex signaling pathways.
2. Mechanism of Transcriptional Activation of Hepatic Genes during Liver Development
Transcriptional activation of genes during development is mediated by several key hepatic regulators, which act in concert with specific signaling pathways to establish expression profiles that define differentiation-specific cellular states. Accumulating evidence suggests that regulatory regions (enhancers and promoters) of tissue-specific genes often reside in compacted genomic regions that cannot be accessed by transcription factors, thus acting as a barrier to transcription. Initial gene activation requires a defined sequence of transcription factor–DNA interactions and chromatin transitions, which can cope with the structural obstacle of chromatin condensation. This has become the prevailing view, following the discovery of a special class of transcription factors, now known as pioneer factors. These pioneer factors possess the ability to bind their recognition sequence when embedded into a highly condensed chromatin state.
Pioneer factors were discovered in an attempt to uncover the first transcription factor that binds to the enhancer of the liver-specific albumin gene during embryogenesis. In vivo footprinting studies in an enhancer of the serum albumin gene showed that FoxA and GATA factors occupied their target sites both in pluripotent endoderm, where the Alb gene was silent, and in the nascent liver bud, where the Alb gene was expressed [1][2]. When assessing the binding affinity of these factors by in vitro experiments, it was observed that both were able to bind to compacted chromatin and open the local nucleosomal domain without the requirement for ATP or ATP-dependent chromatin remodelers. Thus, FoxA1 and Gata4 have the ability to bind to heterochromatin and occupy their target sequences prior to transcriptional activation. Because these binding events define the initiating step in developmental gene activation, FoxA1 and Gata4 proteins were named “pioneer” transcription factors. So far, studies indicate that pioneer factors have four distinct features: a. they bind to their targets embedded in a closed chromatin state, b. they increase the accessibility in the target region for other proteins, c. they regulate cell programming, and d. they establish a stable epigenetic memory mechanism [3][4].
Gene activation during development includes several steps. Initially, pioneer factors scan the genome and bind to particular regions as they encounter their binding sites [4]. There are many potential binding sites for pioneer factors, but only a subset of these sites are occupied. This selective genomic occupancy is cell type-dependent and is regulated by cell type-specific co-factors, the state of the chromatin domains, and various signaling pathways [5][6][7][8][9][10][11][12][13]. The initial binding in the closed, silent chromatin is weak but appears to be rapid [14]. This is followed by a slower process in which the local chromatin is re-organized and becomes more accessible. Pioneer factors are necessary for the kick-starting of changes in the chromatin, but they are unable to induce transcription on their own accord. For this to take place, other components of the transcription apparatus such as other transcription factors, chromatin modifiers, and nucleosome remodelers must cooperate with the pioneer factors [15] to modify nucleosome structure and facilitate preinitiation complex formation for an efficient RNA Polymerase-II loading [4][16][17] ( Figure 1 ).
3. Developmental Bookmarking by Pioneer and Non-Pioneer Transcription Factors
Pioneer factors act as priming factors to establish the transcriptional competence of their target genes during development, but their binding is not accompanied by immediate transcription activation. This priming activity can be attributed to their potential role as “bookmarking” factors. In other words, following initial chromatin opening, pioneer factors remain associated with the regulatory regions and keep the loci competent for the future assembly of an active preinitiation complex. During this time, other factors may be recruited to the now accessible regulatory regions and build a preinitiation complex.
A recent study has shown that the recruitment of two prominent hepatic regulators, HNF4α and C/EBPα, similarly to FoxA1, is not linked to concomitant gene activation during development [18]. The time between transcription factor binding and gene activation ranges from a few days to weeks. This is considered quite a substantial amount of time in mouse development. What happens during this time? Is bookmarking a “static” process, where pioneer and non-pioneer factors simply mark the locus to prevent “re-compaction”? Does the time difference between transcription factor binding and developmental gene activation simply reflect the lack of availability of some specific activating signals, which influence the recruitment or activation of additional factors required for transcription initiation?
Insights into the above mentioned questions were provided by studying the dynamics of transcription factor recruitment and chromatin structure changes during developmental gene activation. It was observed that dynamic binding events, i.e., the transient binding of transcription factors, without gene activation is the most common phenomenon during development. The stable and transient association of transcription factors with different cis-regulatory elements in promoter and enhancer regions facilitates the recruitment of chromatin remodelers and the generation of active chromatin configurations. The length of time during which such dynamic interactions take place in a continuous fashion allows for the cumulative increase in histone modifications characteristic of active enhancers and the progressive expansion of stably open chromatin domains. In this way, bookmarking is part of a highly dynamic developmental maturation process during which regulatory regions are prepared for the acquisition of an optimal configuration that supports an efficient and stable transcription (Figure 1).
The model above was supported by the analyses of mice that were deficient in the bookmarking factors HNF4α or C/EBPα. In both cases, a significant deregulation of transcription of most early-bound hepatic genes was observed in parallel to the blocking of acquisition in active chromatin states and the reciprocal accumulation of repressive histone modification marks [18].
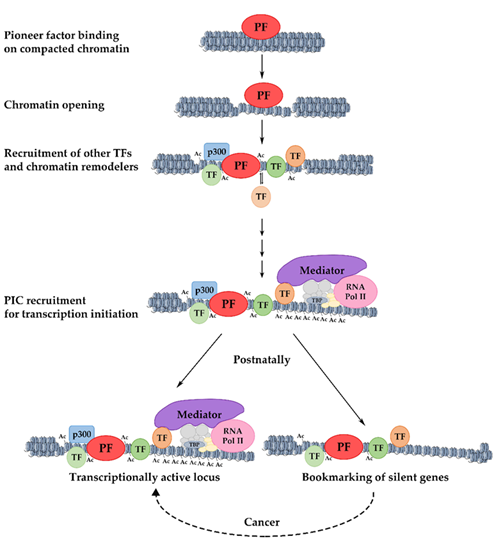
Figure 1. Mechanism of pioneer factor activity, transcriptional activation, and bookmarking. The initial binding of a pioneer factor to its target sites occurs in highly condensed chromatin and results in increased chromatin accessibility. The progressive recruitment of chromatin modifiers and the stable or transient binding of other transcription factors lead to the gradual deposition of activating histone modifications and the broadening of active chromatin domains. The resulting permissive chromatin state facilitates the assembly of the pre-initiation complex (PIC) and promotes transcriptional initiation. Loci that are postnatally silenced retain transcription factors on their promoters, keeping them competent for re-activation under certain conditions. PF: pioneer factor; TF: transcription factor.
4. Maintenance of Stable Hepatic Gene Expression Patterns
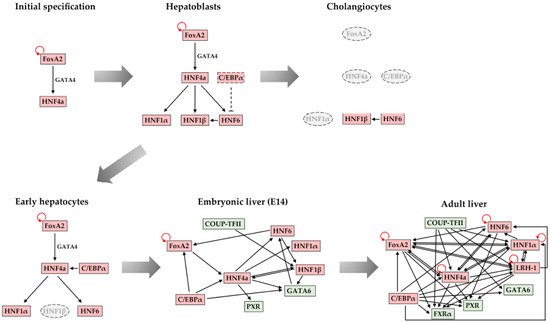
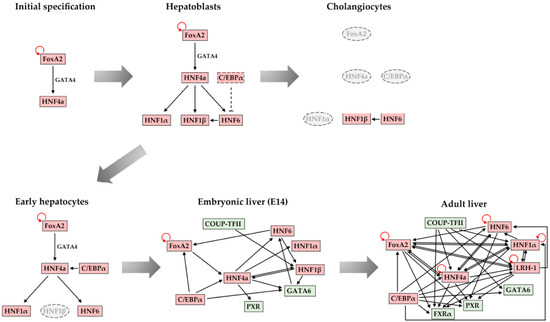
5. Conclusions and Future Perspectives
This entry is adapted from the peer-reviewed paper 10.3390/cells10082026
References
- Gualdi, R.; Bossard, P.; Zheng, M.; Hamada, Y.; Coleman, J.R.; Zaret, K.S. Hepatic specification of the gut endoderm in vitro: Cell signaling and transcriptional control. Genes Dev. 1996, 10, 1670–1682.
- Bossard, P.; Zaret, K.S. GATA transcription factors as potentiators of gut endoderm differentiation. Development 1998, 125, 4909–4917.
- Iwafuchi-Doi, M.; Zaret, K.S. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014, 28, 2679–2692.
- Mayran, A.; Drouin, J. Pioneer transcription factors shape the epigenetic landscape. J. Biol. Chem. 2018, 293, 13795–13804.
- Lupien, M.; Eeckhoute, J.; Meyer, C.A.; Wang, Q.; Zhang, Y.; Li, W.; Carroll, J.S.; Liu, X.S.; Brown, M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 2008, 132, 958–970.
- Zaret, K.S.; Mango, S.E. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr. Opin. Genet. Dev. 2016, 37, 76–81.
- Hurtado, A.; Holmes, K.A.; Ross-Innes, C.S.; Schmidt, D.; Carroll, J.S. FOXA1 is a critical determinant of Estrogen Receptor function and endocrine response. Nat. Genet. 2011, 43, 27–33.
- Chen, J.; Zhang, Z.; Li, L.; Chen, B.C.; Revyakin, A.; Hajj, B.; Legant, W.; Dahan, M.; Lionnet, T.; Betzig, E.; et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 2014, 156, 1274–1285.
- Swinstead, E.E.; Miranda, T.B.; Paakinaho, V.; Baek, S.; Goldstein, I.; Hawkins, M.; Karpova, T.S.; Ball, D.; Mazza, D.; Lavis, L.D.; et al. Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell 2016, 165, 593–605.
- Liu, Z.; Kraus, W.L. Catalytic-independent functions of PARP-1 determine Sox2 pioneer activity at intractable genomic loci. Mol. Cell 2017, 65, 589–603.
- Franco, H.L.; Nagari, A.; Kraus, W.L. TNFα signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol. Cell 2015, 58, 21–34.
- Soufi, A.; Donahue, G.; Zaret, K.S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 2012, 151, 994–1004.
- Soufi, A.; Fernandez Garcia, M.; Jaroszewicz, A.; Osman, N.; Pellegrini, M.; Zaret, K.S. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 2015, 161, 555–568.
- Mayran, A.; Khetchoumian, K.; Hariri, F.; Pastinen, T.; Gauthier, Y.; Balsalobre, A.; Drouin, J. Pioneer factor Pax7 deploys a stable enhancer repertoire for specification of cell fate. Nat. Genet. 2018, 50, 259–269.
- Iwafuchi-Doi, M.; Zaret, K.S. Cell fate control by pioneer transcription factors. Development 2016, 143, 1833–1837.
- Soutoglou, E.; Papafotiou, G.; Katrakili, N.; Talianidis, I. Transcriptional activation by hepatocyte nuclear factor-1 requires synergism between multiple coactivator proteins. J. Biol. Chem. 2000, 275, 12515–12520.
- Soutoglou, E.; Viollet, B.; Vaxillaire, M.; Yaniv, M.; Pontoglio, M.; Talianidis, I. Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J. 2001, 20, 1984–1992.
- Karagianni, P.; Moulos, P.; Schmidt, D.; Odom, D.T.; Talianidis, I. Bookmarking by non-pioneer transcription factors during liver development establishes competence for future gene activation. Cell Rep. 2020, 30, 1319–1328.
- Reizel, Y.; Morgan, A.; Gao, L.; Lan, Y.; Manduchi, E.; Waite, E.L.; Wang, A.W.; Wells, A.; Kaestner, K.H. Collapse of the hepatic gene regulatory network in the absence of FoxA factors. Genes Dev. 2020, 34, 1039–1050.
- Si-Tayeb, K.; Lemaigre, F.P.; Duncan, S.A. Organogenesis and development of the liver. Dev. Cell 2010, 18, 175–189.
- Lemaigre, F.P. Mechanisms of Liver Development: Concepts for Understanding Liver Disorders and Design of Novel Therapies. Gastroenterology 2009, 137, 62–79.
- Kyrmizi, I.; Hatzis, P.; Katrakili, N.; Tronche, F.; Gonzalez, F.J.; Talianidis, I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev. 2006, 20, 2293–2305.
