Oxidative stress biomarkers can be used to detect oxidative damages occurring concomitantly in many cellular structures, which may cause a deterioration of function, including apoptosis and necrosis. There is an intimate relationship between oxidative stress, inflammation, and functional impairment, resulting in various diseases affecting the entire human body.
- oxidative stress
- biomarkers
- antioxidants
- lipid peroxidation
- protein peroxidation
- DNA peroxidation
- signaling pathway
- kidney
- renal transplantation
- ischemia-reperfusion injury
1. Introduction
Reactive oxygen species are compounds that are difficult to measure when assessing oxidative stress, primarily due to the very short half-life, so they hardly play the role of biomarkers. However, if ROS combines with a particular biological molecule, it leaves a unique chemical “fingerprint”. Biomarkers obtained that way can be used to evaluate oxidative damage or the effects of antioxidants, including therapeutic agents. The core criterion for the biomarker is its role in the prediction of the later development of disease. Moreover, important technical criteria of a biomarker are it should detect a major part of total ongoing oxidative damage in vivo, should provide coherent laboratory assays, results should not vary under the same conditions, should be stable during storage, must employ chemically robust measurement technology, and must not be confounded by diet [1]. There is no ideal biomarker, yet many provide sufficient accuracy. ROS, as highly reactive substances, interact with the environment in vivo, involving and stimulating various endogenous mechanisms as well as react with numerous molecules, leaving a mentioned fingerprint, which becomes the point of interest in specific evaluations. ROS, reactions, and essential antioxidants were presented in Figure 1.
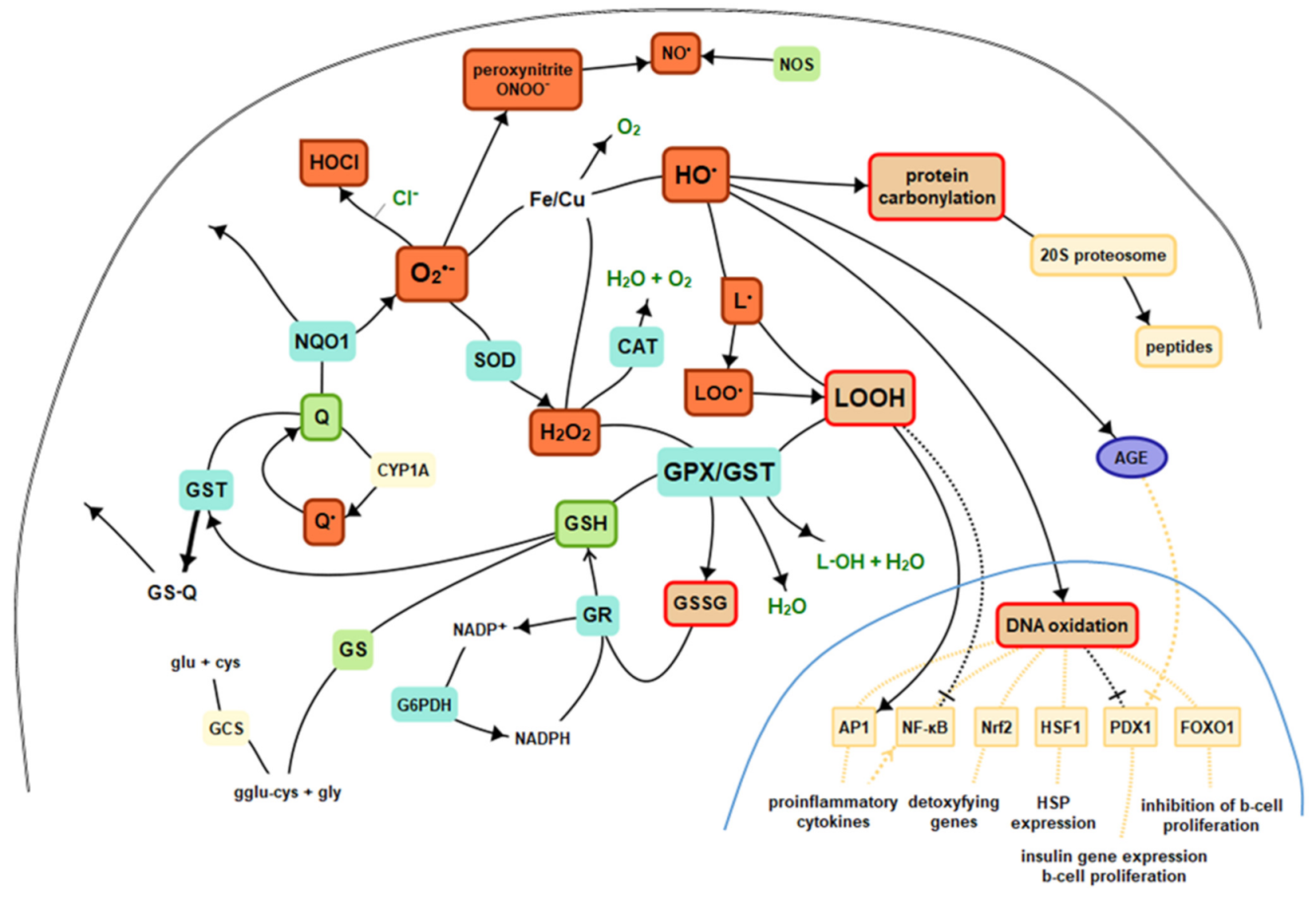 Figure 1. Basics of intracellular antioxidant mechanisms and nuclear signaling. Legend: CAT—catalase, GPx—glutathione peroxidase, GR—glutathione reductase, GST—glutathione S-transferases, GSH—reduced glutathione, GSSG—oxidized glutathione, Q—coenzyme Q10, GS—glutathione synthetase, HO·—hydroxyl radical, H2O2—hydrogen peroxide, L·—alkyl radical, LO·—alkoxy radical, LOO·—peroxy radical, HOCl—hypochlorous acid, NO—nitric oxide, NOS—nitric oxide synthase, NQO1—NADPH-quinone oxidoreductase-1, O2—molecular oxygen, O2•—superoxide, SOD—superoxide dismutase, AGE—advanced glycation end products, NF-κB—nuclear factor kappa-light-chain-enhancer of activated B cells, AP1—activator protein 1, Nrf2—nuclear factor-erythroid 2-related factor 2, HSF1—heat shock factor 1, PDX1—insulin promoter factor 1, FoxO—forkhead transcription factor O.
Figure 1. Basics of intracellular antioxidant mechanisms and nuclear signaling. Legend: CAT—catalase, GPx—glutathione peroxidase, GR—glutathione reductase, GST—glutathione S-transferases, GSH—reduced glutathione, GSSG—oxidized glutathione, Q—coenzyme Q10, GS—glutathione synthetase, HO·—hydroxyl radical, H2O2—hydrogen peroxide, L·—alkyl radical, LO·—alkoxy radical, LOO·—peroxy radical, HOCl—hypochlorous acid, NO—nitric oxide, NOS—nitric oxide synthase, NQO1—NADPH-quinone oxidoreductase-1, O2—molecular oxygen, O2•—superoxide, SOD—superoxide dismutase, AGE—advanced glycation end products, NF-κB—nuclear factor kappa-light-chain-enhancer of activated B cells, AP1—activator protein 1, Nrf2—nuclear factor-erythroid 2-related factor 2, HSF1—heat shock factor 1, PDX1—insulin promoter factor 1, FoxO—forkhead transcription factor O.
2. Oxidative Stress Biomarkers
2.1. Endogenous Antioxidants
| Biomarker | Description | General | Transplantation |
|---|---|---|---|
| CAT catalase |
tetramer, enzyme, biomarker, catalyzes the decomposition of hydrogen peroxide to water and oxygen, has one of the highest turnover numbers of all enzymes, first noticed in 1818 |  1068 |
 356 |
| SOD superoxide dismutase |
enzyme, biomarker, metalloprotein connected in humans with Zn/Cu or Mn, discovered in 1968 |  2117 |
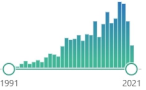 715 |
| GPx glutathione peroxidase |
selenium-containing enzyme family, several isozymes are encoded by different genes, biomarker, the protective system depends heavily on the presence of selenium, discovered in 1957 | 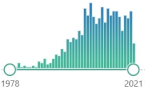 763 |
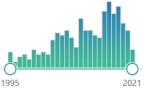 296 |
| GST glutathione S-transferase |
family of metabolic isoenzymes, biomarker, catalyze the conjugation of the reduced form of glutathione (GSH) to xenobiotic substrates, nomenclature first proposed in 1992 | 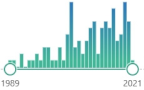 104 |
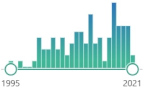 95 |
| GR glutathione reductase |
homodimer disulfide oxidoreductase enzyme encoded by the GSR gene, biomarker, catalyzes GSSH and regenerates it to GSH, involves NADPH and FAD binding domains, first purified in 1955 | 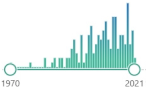 159 |
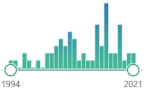 63 |
| HO-1 heme oxygenase 1 |
an enzyme, mediates the first step of heme catabolism, cleaves heme to biliverdin, carbon monoxide, and ferrous iron, encoded by HMOX1 gene induced in oxidative stress and inflammation, first characterized in 1962 | 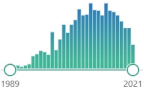 1457 |
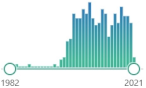 404 |
| NQO1 NADPH-quinone oxidoreductase-1 |
protein homodimer, detoxifying enzyme, present in cytosol, performs a two-electron reduction of quinones to hydroquinones and of other redox dyes without the formation of ROS, | 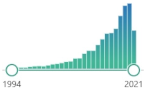 1837 |
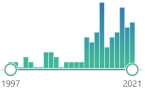 86 |
| GSH glutathione |
L-γ-glutamyl-L-cysteinyl-glycine, non-enzymatic, biomarker, reduced form (GSH) and glutathione disulfide (GSSG)—primary redox couple in animal cells | 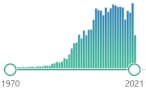 5118 |
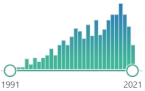 798 |
| coenzyme Q ubiquinone |
benzoquinone derivate, localized in the mitochondrial respiratory chain and other internal membranes, coenzyme, first isolate was in 1950 in the lining of a horse’s gut | 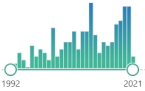 185 |
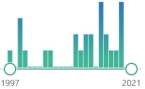 22 |
| ALA α-lipoic acid |
delivers antioxidant activity in nonpolar and polar mediums, effective in recharging enzymes in the mitochondria, might counteract NF-κB activation | 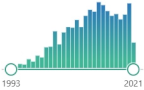 1430 |
 62 |
| BR bilirubin |
product of heme degradation works as an antioxidant in cycle BR-biliverdin, relevantly documented in 1827 |  1941 |
 64 |
| ferritin | intracellular globular iron-binding protein, small amounts are secreted into the serum, prevents ROS generation via the Fenton reaction by binding iron |  1678 |
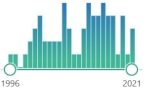 60 |
2.2. Lipid Peroxidation (LPO)
| Biomarker | Description | General | Transplantation |
|---|---|---|---|
| TBARS TBA-reactive substances |
the oldest and one of the most widely used nonspecific by-products of lipid peroxidation, reacts with thiobarbituric acid (TBA), forming a pink chromogen (TBARS) measured at 532–535 nm | 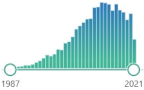 7431 |
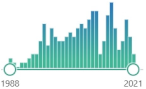 200 |
| MDA malondialdehyde |
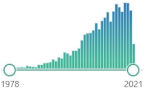
CH2(CHO)2, colorless liquid, highly reactive, a product of LPO of polyunsaturated fatty acids, form covalent protein adducts referred to as advanced lipoxidation end-products (ALE), in analogy to advanced glycation end-products (AGE) | 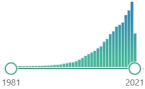 30,269 |
 2808 |
| 4-HNE 4-hydroxynonenal |
α,β-unsaturated hydroxyalkenal, produced by lipid peroxidation (arachidonic or linoleic groups) in cells in higher quantities during oxidative stress, possible role in cell signal transduction, first reported in 1991 [2], they can also come from omega-3 fatty acids | 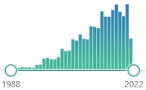 2418 |
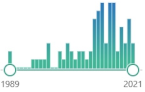 73 |
| ACR acrolein (propenal) |
the simplest unsaturated aldehyde, named and characterized in 1839, electrophilic, reactive and toxic, contact herbicide to weeds, present in tobacco smoke increases the risk of cancer, produced during cyclophosphamide treatment | 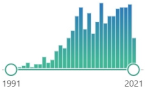 547 |
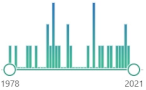 29 |
| F2-isoprostanes | prostaglandin-like compounds formed in vivo from the free radical-catalyzed peroxidation of arachidonic acid, discovered in 1990, | 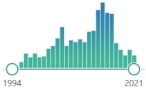 623 |
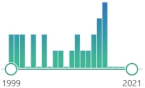 22 |
| F4-isoprostanes | prostaglandin-like compounds formed in vivo from the free radical-catalyzed peroxidation of docosahexaenoic acid, potent biological activity as anti-inflammatory mediators |  3 |
0 |
| CRA crotonaldehyde |
CH3CH, representative carcinogenic aldehyde formed endogenously through lipid peroxidation, CRA is a highly reactive aldehyde and reacts with a lysine residue in the protein, reaction with CRA and lysine residue leads to the formation of numerous numbers of adducts | 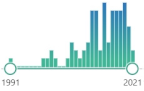 74 |
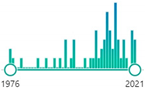 53 |
| HHE 4-hydroxy-trans-2-hexenal |
oxygenated α,β-unsaturated aldehyde, other coming from omega-3 fatty acids: 4-oxo-trans-2-nonenal, 4-hydroperoxy-trans-2-nonenal, and 4,5-epoxy-trans-2-decenal | 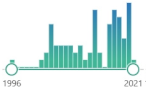 64 |
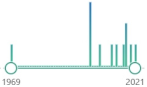 11 |
| 7KC 7-ketocholesterol (7-oxocholesterol) |
toxic oxysterol, produced from oxidized cholesterol, induces: NOX, pro-inflammatory cytokines and TNF-α | 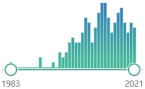 162 |
 6 |
2.3. Protein Oxidation
| Biomarker | Description | General | Transplantation |
|---|---|---|---|
| DiBrY dibromotyrosine |
product of the reaction of hypobromous acid (HOBr) from hydrogen peroxide (H2O2) and bromide ion (Br–), detected by anti-dibromo-tyrosine [DiBrY], mAb (3A5) JAI-MBY-020P |  52 |
5 1951, 1989, 1995, 1999, 2018 |
| DiY/DT dityrosine (bityrosine) |
biphenyl compound comprising two tyrosine residues linked at carbon-3 of their benzene rings |  1014 |
6 2001, 2005, 2010, 2012, 2020(2) |
| m-Tyrosine o-Tyrosine |
abnormal tyrosine isomers, derive from oxidation of the benzyl ring of the phenylalanine by hydroxyl radical, adversely affect cells and tissues |  62 |
4 2003, 2007, 2008, 2012 |
| NY 3-nitrotyrosine |
specific marker of attack of peroxynitrite (ONOO–) upon proteins, measured by immunostaining, HPLC, and MS in human tissues | 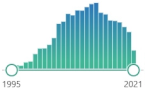 3554 |
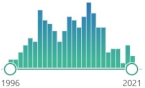 201 |
| protein carbonyls | measurement of protein CO groups after their derivatization with DNPH is the most widely utilized measure of protein oxidation | 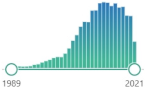 9122 |
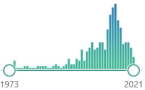 325 |
2.4. Nucleic Acid Oxidation
| Biomarker | Description | General | Transplantation |
|---|---|---|---|
| 8OHdG 8-hydroxy-2′ -deoxyguanosine |
oxidized derivative of deoxyguanosine, major products of DNA oxidation, increased levels are found during carcinogenesis, increases with age, linked to the enzyme OGG1 and transcription factor NFκB | 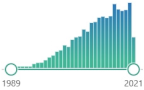 5189 |
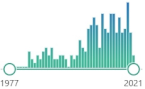 212 |
| 8-oxo-Gua 8-hydroxyguanine |
one of the most common DNA lesions resulting from reactive oxygen species, modifying guanine, and can result in a mismatched pairing with adenine resulting in G to T and C to A → mutation | 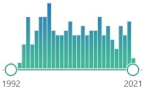 199 |
0 |
3. Conclusions
This entry is adapted from the peer-reviewed paper 10.3390/ijms22158010
References
- Halliwell, B.; Whiteman, M. Measuring reactive species and oxidative damage in vivo and in cell culture: How should you do it and what do the results mean? Br. J. Pharm. 2004, 142, 231–255.
- Esterbauer, H.; Schaur, R.J.; Zollner, H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free. Radic. Biol. Med. 1991, 11, 81–128.
- Dizdaroglu, M.; Jaruga, P.; Birincioglu, M.; Rodriguez, H. Free radical-induced damage to DNA: Mechanisms and measurement. Free. Radic. Biol. Med. 2002, 32, 1102–1115.
- Chiorcea-Paquim, A.-M.; Oliveira-Brett, A.M. Nanostructured material-based electrochemical sensing of oxidative DNA damage biomarkers 8-oxoguanine and 8-oxodeoxyguanosine: A comprehensive review. Mikrochim. Acta 2021, 188, 58.
