Molecularly imprinted polymers (MIP) are obtained by initiating the polymerization of functional monomers surrounding the template molecule in the presence of crosslinkers and porogens. Usually the best adsorption performance can be obtained by optimizing the polymerization conditions, but the process is time-consuming and labor-intensive. At the same time, the use of a large number of organic reagents in the process of experimental optimization also limits the development and promotion of molecular imprinting technology. Theoretical calculation based on calculation simulation and intermolecular force is an effective method to solve this problem because it is convenient, versatile, environmentally friendly and low in price. It is not affected by the space environment, and the calculation efficiency is high. This article reviews the theoretical simulation calculation methods used in the preparation of MIPs in recent years.
- computational simulation
- molecularly imprinted polymers
- intermolecular interaction
一、简介[ 1 ] [ 2 ] [ 3 ] [ 4 ]
Molecularly imprinted polymers (MIPs) are porous materials with specific recognition capacity towards the template molecule, which are obtained by self-assembly of template molecules and functional monomers in a porogen, and then polymerization is initiated in the presence of a cross-linking agent. The process of preparing MIPs is outlined in Figure 1. When the template molecule interacts with the functional monomer, the imprinting site is memorized through multiple action effects and fixed through the polymerization process. After the template is removed, the adsorption cavity complementary in shape and structure to the template molecule is left in the polymer matrix, which can selectively recognize the target molecule. Molecular imprinting technology originated from antibody immunology, that is, the specific combination of “lock and key” between antibody and antigen [1]. In 1973, Wulff [2] prepared organic MIPs for the first time. Since then, MIPs have attracted widespread attention. At present, MIPs, as a kind of intelligent adsorption material, are widely used in various fields, such as chromatographic separation [3], solid phase extraction [4–6], sensors [7–9], and biomedicine [10,11]. In the past two decades, great progress in MIPs has been achieved (Figure 2). A variety of novel and interesting imprinted polymers, including supramolecular imprinted polymers [12,13], multitemplate imprinted polymers [14,15], multifunctional monomer imprinted polymers [16,17], dummy template imprinted polymers [18,19], and chiral recognition polymers [20,21], have been developed. In fact, synthesis parameters have been obtained through experimental optimization in most cases. Finding complex and cumbersome conditions is time consuming and laborious. Moreover, numerous organic reagents are used. These factors severely restrict the application and promotion of molecular imprinting technology.
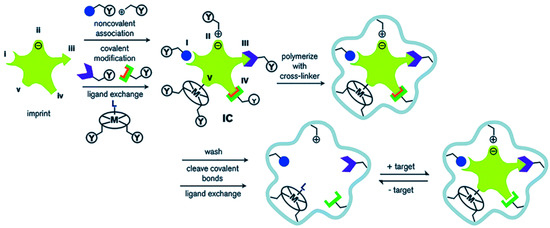
Figure 1. Schematic diagram of the molecular imprinting process: (I) non-covalent, (II) electrostatic/ionic, (III) covalent, (IV) semi-covalent, and (V) coordination to a metal center (Reprinted with permission from [22]. Copyright 2014 Royal Society of Chemistry).
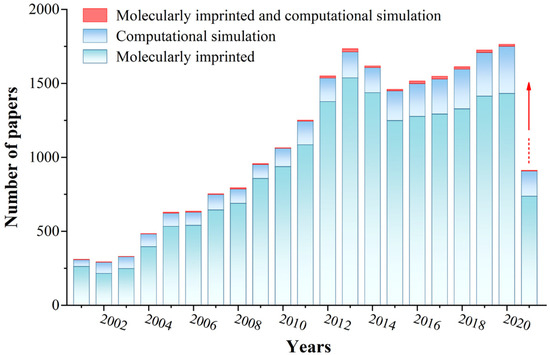
Figure 2. The literature statistics of MIPs and computational simulation. (Database: Scifinder; Search keywords: molecularly imprinted, computational simulation, molecularly imprinted and computational simulation, respectively. Search time: 13 June 2021).
2.Theoretical Methods of Computational Simulation for MIPs
The methods used in the theoretical calculation and simulation of various MIP designs are molecular mechanics (MM), molecular dynamics (MD), and quantum mechanics (QM). The computational cost of MM optimization is considerably lower than that of QM, and thus it is orders of magnitude faster than the latter. However, the accuracy of MM results is limited by simplified calculation models, which allow the reduction in calculation costs. The QM approach can better solve the problem of choosing the appropriate initial direction of interacting molecules because it is more accurate than the other methods. However, the computational complexity of the QM approach exponentially increases as the number of molecules involved in the calculation system increases. The MD method can effectively address this problem. When simulating the dynamic process of the interaction between molecules, changes in the molecule itself are often not considered, thereby making the calculation of the simulation method more efficient. Therefore, the MD method is most widely used when numerous molecules are involved in designing MIPs, such as in optimizing the ratio of template, monomer, and cross-linking agent. The application of MM, MD, and QM methods in MIP simulation is given in Table 1.
Table 1. Theoretical simulation calculation methods for the design of MIPs.
|
Simulation Method |
Template |
Force Field/Method |
Software |
MIPs Design |
|---|---|---|---|---|
|
Molecular mechanics (MM) |
Myoglobin [29] |
OPLS3 |
Prime |
Screening functional monomers |
|
Morphine [30] |
CHARMM and MMFF94 |
Discovery Studio |
Template-monomer ratio |
|
|
Metolachlor deschloro [31], metsulfuron-methyl [32] |
AMBER MM |
SPSS Statistics |
Screening functional monomers/template-monomer ratio |
|
|
Norfloxacin [33] |
MMFF94X |
Discovery Studio |
Screening functional monomers/template-monomer ratio |
|
|
Molecular dynamics (MD) |
Curcumin [34], fenthion [35], N-3-oxo-dodecanoyl-L-homoserine lactone [36], methidathion [37], endotoxins [38], phosmet insecticide [39], cocaine [40], methyl parathion [41], aflatoxin B1 [42] |
Tripos |
SYBYL |
Screening functional monomers/template-monomer ratio |
|
Bisphenol A [43], carbamazepine [44], phthalates [45], norfloxacin [46], sulfamethoxazole [47] |
COMPASS |
Materials Studio/accelrys.com |
Screening functional monomers/template-monomer ratio |
|
|
Thiamethoxam [48] |
AMBER |
Gaussian |
Template-monomer ratio and solvent |
|
|
Rhodamine B [49] |
GROMOS |
GROMACS |
Template-monomer ratio and solvent |
|
|
Quantum mechanics (QM) |
Vancomycin [50], primaquine [51], tramadol [52], thiamethoxam [48], clenbuterol [53], sulfadimidine [54], bilobalide [55], chloramphenicol [56], paclitaxel [57], acetamiprid [58], acetazolamide [59], lamotrigine [60], cyanazine [61], 3-methylindole [62], polybrominated diphenyl ethers [63], pirimicarb [64], metoprolol [65], ciprofloxacin or norfloxacin [66] |
DFT |
Gaussian |
Screening functional monomers/template-monomer ratio |
|
Aspartame [67], pinacolyl methylphosphonate [68], metolachlor deschloro [31], metsulfuron-methyl [32], thiocarbohydrazide [69] |
Semiempirical method |
Spartan/SPSS Statistics |
Screening functional monomers/template-monomer ratio |
|
|
Benzo[a]pyrene [70], tryptophan [71], furosemide [72], buprenorphine [73], hydroxyzine and cetirizine [74], atenolol [75], diazepam [76], metolachlor deschloro [31], metsulfuron-methyl [32], allopurinol [77], methadone [78], clonazepam [79], theophylline [80], ametryn [81], mosapride citrate [82], baicalein [83] |
Ab initio |
HyperChem/Gaussian/AutoDockTools/SPSS Statistics |
Screening functional monomers/template-monomer ratio |
|
|
|
3.Computational Simulation and Design of New MIPs
The application of theoretical calculations in designing MIPs is primarily achieved by theoretical simulations and selection of appropriate functional monomers, template molecules, crosslinkers, and their ratios. The binding energy (i.e., electronic interaction energy) between the template molecule and the functional monomer can be simulated and calculated provided that the binding energy between the template molecule and the functional monomer is high, indicating that the corresponding MIPs have excellent selectivity and adsorption performance. In addition, the ratio of the molecular and monomer system is closely related to the imprint factor of MIPs. In general, this ratio is calculated and optimized by performing the computational simulation in a vacuum environment to obtain the Eqaution (1) for the binding energy between the template molecule and the functional monomer.
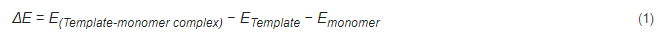
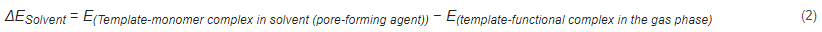
where ΔESolvent is the energy difference between a template molecule and a functional monomer in solution and in a vacuum environment. A weak influence of the solution on noncovalent interactions during molecular fingerprint polymerization results in a small energy difference value, suggesting that the solvent is the best polymerization solvent for obtaining molecular fingerprint polymers [99,100].
The primary factor in MIP imprinting polymerization is the strong bonding force between the template and the functional monomer. Therefore, choosing the right functional monomer is a key factor in designing MIPs. An MIP can be reasonably designed by applying the DFT method in selecting the monomer with the best interaction with 2-isopropoxyphenol; it can be combined with the PM3.5 method to optimize the template-to-monomer ratio [101]. Quantum calculations were performed using the Spartan software, and the complexes’ binding energy can be obtained to evaluate their stability. Pyrrole had been selected as the best functional monomer for designing 2-isopropoxyphenol MIPs. PM3 and DFT calculation methods were also used to simulate and calculate the monomers with the strongest interaction with disulfoton [102], chlorogenic acid [103], and amoxicillin [104], as well as the best ratio between the two. This method can be further used to calculate the solution energies of baicalein and acrylamide complexes in different solvents to screen the best polymerization solvent [105].
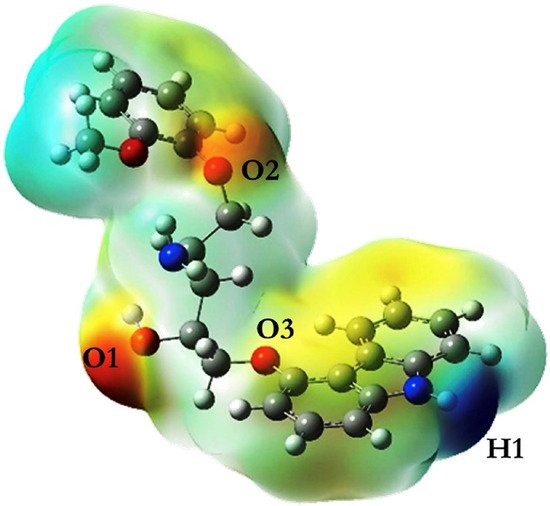
The strongest interaction site can be further located by obtaining the electrostatic potential map on the surface of the template molecule via the DFT method [106]. Figure 4 shows the electrostatic charge distribution of carvedilol after the geometry was optimized. The hydrogen bonding sites between carvedilol and functional monomer evidently appear in the red, yellow, and blue regions, which were O1, O2, O3, and H1. According to the quantitative information of the electrostatic map, each functional monomer undergoes hydrogen bonding at the four interaction sites in sequence to form hydrogen bonds; thus, the ratio of template and monomer complexes were 1:1 and 1:2, and 1:3 and 1:4. When the functional monomer is methacrylic acid and the template is combined with the monomer at a ratio of 1:4, a stable complex can be formed. The DFT method had also been adopted to study the interaction between p-nitrophenol and β-cyclodextrin [12].
4.Computational Simulation and MIP Identification Mechanism
The selective mechanism of ciprofloxacin-imprinted membrane was also further explained through molecular simulations [127]. The binding energy of the interaction between the functional monomer and ciprofloxacin and its structural analogs, including norfloxacin hydrochloride, enrofloxacin hydrochloride and ofloxacin hydrochloride, was obtained through molecular simulation calculations. Kinetic simulations had also been performed using GROMACS software. The parameters of bond, angle, dihedral angle, and Lennard–Jones interaction had been directly taken from the GAFF force field. Part of the charge was obtained using the restricted electrostatic potential method at the theoretical level of B3LYP/6-31+G (d, p). The recombination ability of the imprinted site of ciprofloxacin was dominated by hydrogen bond interactions, whereas its structural analogs were dominated by van der Waals interaction. Thus, strong hydrogen bond interactions led to a high tendency for the imprinted site of ciprofloxacin to recombine with the template molecule. Theoretical simulations of the recombination mechanism and selective permeation experiments mutually confirmed the superior selectivity of ciprofloxacin-imprinted membranes. Zhang [128] further explained the specific selective recognition mechanism of molecularly imprinted nanocomposite membranes for artemisinin by dynamic simulations. A comparison of the binding energy of the imprinted membrane with artemisinin and its structural analogs shows that the strong interaction between artemisinin and the imprinted polymer matrix contributes to its large adsorption capacity and high selectivity. A similar DFT method has been used to explain the selective recognition mechanism of the alternative template N-(4-isopropylphenyl)-N′-butyleneurea MIP to phenylurea herbicides [129]. This method also explained the mechanism of experimentally preferred dummy template imprinted polymer [130] and the strength of the bonding force of chiral naproxen MIP [21] at the molecular level. These observations provide a theoretical basis to explain the experimental results from the perspective of intermolecular interactions.
Yang [61] performed molecular simulation to reveal the essential reason for the difference between single-template and double-template MIP stirring bars in their ability to recognize target analytes by using the YASARA software to study the recognition mechanism. The 3D shape and size of the imprinted cavity in the MIPs are the corresponding template molecules. Given that the dual-template MIP contains imprinted cavities of the two template molecules, it had a fairly high recovery for nine fluoroquinolones, and the simulation results are consistent with the experimental findings. However, the influence of template–template interactions on the performance of multitemplate MIPs has been further verified via the DFT method [62]. The results of both theoretical simulations and experiments indicated that the interaction between more template molecules affects the formation of specific recognition sites and even reduces the formation of effective imprinting sites.
5. Conclusions and Outlook
Computer molecular modeling technology has been applied to the screening and optimization of molecules in many materials, and it is also a feasible method for preliminary exploration of MIP. Computer simulation reduces the time and reagent-related costs required to obtain the appropriate MIP adsorbent, and significantly reduces the consumption of organic solvents. In addition, it can explain the specific recognition mechanism of imprinted materials at the molecular level. For all the above reasons, the use of computer molecular simulation to design MIP adsorbents in analytical practice not only conforms to the principles of green analytical chemistry, but also explains the nature of MIPs binding to target molecules from the intermolecular forces. The QM method, compared with other methods, can ensure more accurate simulation results in the MIP system dominated by non-bonding interaction, because the smallest structural unit electron was studied and the quantum effect was considered in the method. Therefore, the QM method is also the most widely used in MIP simulation operations. However, in the simulation of macromolecules and polyatomic systems, this method is very time-consuming and even prone to errors. MM and MD are classical mechanics methods. Their smallest structural unit is no longer an electron but an atom. Therefore, the simulation operation complexity of the imprinting system is greatly reduced, and the operation speed is faster. MM method directly utilizes the potential function to study the problem, without considering the kinetic energy and the corresponding structure of the atom. However, the MD method focuses on the movement of atoms in the MIP system and establishes the relationship between temperature and time, which can simulate the imprinting system more realistically, and the simulation results are more representative. In general, the DFT procedure in the QM method was recommended in the MIPs design and mechanism interpretation simulation calculation. However, this also means that the computational complexity of this method increases dramatically for large molecules and systems with a large number of molecules. MD method may be the best solution at this situation, simulated annealing process in particular, which can complete the lowest energy conformation search in a very short time. At present, an increasing number of research have been using multiple calculation methods to achieve complementary advantages when designing and optimizing the experimental parameters of MIPs preparation, so as to ensure more efficient and accurate simulation results. In addition, simulation is also the direction of current efforts. A more realistic simulation environment can make the calculation results accurate and reliable.
This entry is adapted from the peer-reviewed paper 10.3390/polym13162657
References
- Marć, M.; Kupka, T.; Wieczorek, P. P.; Namieśnik, J. Computational modeling of molecularly imprinted polymers as a green approach to the development of novel analytical sorbents. TrAC Trends in Analytical Chemistry 2018, 98, 64–78.
- Paredes-Ramos, M.; Bates, F.; Rodríguez-González, I.; López-Vilariño, J. M. Computational approximations of molecularly imprinted polymers with sulphur based monomers for biological purposes. Materials Today Communications 2019, 20, 100526.
- Cowen, T.; Karim, K.; Piletsky, S. Computational approaches in the design of synthetic receptors – A re-view. Analytica Chimica Acta 2016, 936, 62–74.
- Khan, M. S.; Pal, S.; Krupadam, R. J. Computational strategies for understanding the nature of interaction in dioxin imprinted nanoporous trappers. Journal of Molecular Recognition 2015, 28, 427–437.
