Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
FREM1 (Fras-related extracellular matrix 1) and its splice variant TILRR (Toll-like interleukin-1 receptor regulator) have been identified as integral components of innate immune systems. The potential involvement of FREM1 in HIV-1 (human immunodeficiency virus 1) acquisition was suggested by a genome-wide SNP (single nucleotide polymorphism) analysis of HIV-1 resistant and susceptible sex workers enrolled in the Pumwani sex worker cohort (PSWC) in Nairobi, Kenya.
- FREM1
- TILRR
- inflammation
- HIV-1 acquisition
1. Introduction
FREM1 (Fras-related extracellular matrix 1) plays a critical role in epithelial–mesenchymal interactions [1]. It forms a ternary complex with other integral membrane proteins including FRAS1 (Fraser syndrome 1) and FREM2 [2]. FREM1 functions as an integral component in embryonic development [3], whereas FREM1 variant TILRR (Toll-like interleukin-1 receptor regulator) acts as a modulator of innate immune responses and inflammation [4]. Mutations of FREM1 are associated with multiple phenotypic abnormalities [5]. Because FREM1 is an extracellular matrix (ECM) protein, it may interact with other neighboring ECM partners and promote cell adhesion, migration, proliferation, and differentiation. Studies also showed that TILRR, a splice variant of FREM1, is an IL-1R1 (interleukin 1 receptor, type 1) co-receptor and a highly conserved membrane-associated glycosylated protein [4]. The interactions of TILRR with IL-1-IL-1R1 potentiate the activation of NF-κB transcription factor and inflammatory responses [4][6]. The potential involvement of FREM1 and its variant TILRR in vaginal HIV-1 acquisition was brought about through a genome-wide SNP (single nucleotide polymorphism) analysis. We conducted the study to identify genetic factors associated with the natural resistance to HIV-1 infection observed in a sub-group of female sex workers enrolled in the Pumwani sex worker cohort [7][8][9][10]. The Pumwani sex worker cohort, an open prospective cohort study of sexually transmitted infections (STIs), was established in Nairobi, Kenya in 1985 and the patients enrolled (n > 5000, 1985–2012) in the cohort have been followed biannually since the cohort’s establishment [11][12][13][14]. A sub-group of women enrolled in the cohort remains HIV-1 uninfected despite repeated high-risk exposure through sex work [14]. The genome-wide SNP analysis identified an SNP rs1552896 in FREM1, and its minor allele is enriched in HIV-resistant women [15]. Our subsequent studies showed that FREM1 mRNA is highly expressed in tissues that are relevant to vaginal HIV-1 infection [15]. Furthermore, the FREM1 variant, TILRR, increased the expression of many genes in the IL-1-NF-κB inflammatory signaling pathway [16]. It has been shown that the IL-1-NF-κB signaling pathway functions as an enhancer of HIV-1 acquisition/susceptibility via recruitment and/or activation of HIV-1 target cells [17]. Genital inflammation accompanied by increased activated CD4+T-cells is associated with increased HIV-1 acquisition risk in South African young women [18][19]. Since HIV-1 acquisition is supported by the inflamed genital mucosa where inflammatory mediators fuel the recruitment and activation of HIV-1 target cells [20], it is possible that FREM1 and its variant, TILRR, may be involved in HIV-1 vaginal acquisition/infection through the regulation of inflammatory responses. It is important to emphasize that activated CD4+ T cells support the productive HIV-1 infection [21][22][23]. An inflammatory environment in the female genital tract (FGT) would have a higher amount of activated CD4+ T cells [17][24][25], thus helping HIV-1 to establish and expand infection. Therefore limiting inflammatory responses could reduce the risk of HIV-1 vaginal acquisition. Thus, studying mediators of inflammatory responses, such as TILRR, may help to identify novel HIV-1 prevention targets.
2. Fras-Related Extracellular Matrix 1 (FREM1)
2.1. Molecular Structure of FREM1 and Its Splice Variants
Full-length human FREM1 contains multiple functional domains that interact with extracellular matrix molecules. It has an N-terminal signal sequence, positioned at 1–23 of its amino acid (aa) sequence, and an N-terminal variable region (NV)-containing domain (from aa 24–274) with an Arg-Gly-Asp (RGD) motif (from aa 199–201). Following the NV domain, there are 12 chondroitin sulfate proteoglycan (CSPG) tandem repeats. Each CSPG domain is approximately 120 aa long (from aa 275–1730) [26][27]. At the C-terminal region, it contains a calcium-binding loop of approximately 89 aa long (Calx-β domain, from 1731–1820 aa), and an RGD motif sequence (from 1907–1909 aa). At the end of the C-terminus, it has a unique type C lectin-like (LecC) domain, which is ~131 aa long (from aa 2052–2179) [26]. Additionally, it bears two glycosaminoglycan (GAG) attachment sites located between CSPG10 and CSPG11 domains, and Calx-β and C-terminal RGD motif, which were shown to bind to the IL-1R1 receptor of IL-1-NF-κB signaling pathway (Figure 1A) [16][28].
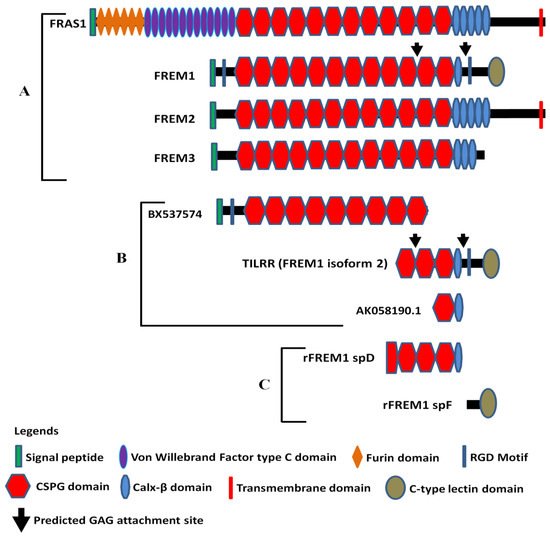
Figure 1. Diagram of FRAS/FREM family proteins, FREM1 splice variants (TILRR and others), and recombinant FREM1 proteins. (A) FRAS/FREM family proteins, including FREM1 (FREM1 isoform 1); (B) TILRR (FREM1 isoform 2) and other truncated variants of FREM1 identified in PBMCs (peripheral blood mononuclear cells) of women enrolled in the Pumwani sex worker cohort; (C) Recombinant FREM1 proteins (rFREM1 spD and rFREM1 spF). RGD, arginine-glycine-aspartic acid; CSPG, chondroitin sulfate proteoglycan; and GAG, glycosaminoglycan. This figure was adapted with permission from Kashem et al. [16], Short et al. [1], and Yuan et al. [28].
The amino acid sequence of N-terminal signal peptide is highly variable between human and mouse FREM1 protein and, therefore, termed as N-terminal variable region-containing domain (NVD) [26]. FREM1 RGD motif sequence is a well-studied cell-adhesive motif and functions as an integrin-binding region. It is recognized by a wide group of integrins, such as αv subunits (αvβ1 and αvβ3) and the β1 subunits (α5β1 and α8β1), which transduce signals to the cells [26]. Twelve CSPG repeats, also known as cadherin repeat-like repetitive elements [3][27] initially identified in NG2 (neural/glial antigen 2) core protein [29], are the hallmark of Fras/FREM family proteins because they are common to all members of the family [3]. Studies have demonstrated that NG2 core proteins (CSPG motifs) frequently interact with an array of extracellular matrix (ECM) proteins, including collagen type II, V, and VI [30][31][32], platelet-derived growth factor (PDGF-AA, a longer isoform of PDGF) [33], and basic fibroblast growth factor (bFGF2) [33]. Thus, it is suggested that FREM1 binds to ECM proteins of basement membranes (BMs) through its repetitive CSPG domains [34] to conduct its biological functions, including cell adhesion, cell proliferation, and migration. The Calx-β domain of FREM1 is a calcium-binding loop of Na+-Ca+ exchange β. The Calx-β domain is characterized by a cytoplasm-localized, calcium chelating, and calcium-binding region of transmembrane proteins, such as integrin β4 [35]. Additionally, the unique feature of FREM1 protein is having a C-terminal LecC domain (CTLD) which is associated with carbohydrate residues of ECM. The other members of Fras/FREM family proteins do not have CTLD [1].
According to AceView [36][37], FREM1 has approximately 15 splice variants. Of them, a full-length isoform 1 encoding the 2179 amino acids long full FREM1 protein (NM_144966.5/NP_659403.4), and a shorter isoform 2 encoding a 715 aa long TILRR protein (NM_001177704.1/NP_001171175.1) were described [34]. The FREM1 isoform 2 (TILRR) has a structurally different N-terminal region and only has 3 CSPG domains with a Calx-β domain and a C- terminal LecC domain [16][28]. Similar to isoform 1, it contains an RGD motif and two GAG attachment sites that are associated with cell adhesion and IL-1β signal transduction, respectively (Figure 1B). In addition, RNA-seq analysis of PBMCs (peripheral blood mononuclear cells) further identified two truncated FREM1 isoforms (BX737574 and AK058190.1) in women of the Pumwani sex worker cohort (PSWC), Nairobi, Kenya (Figure 1B). Unlike FREM1 and its splice variants, the structures of other Fras/FREM-family members are quite different in their N- and C-terminal regions (Figure 1A).
2.2. FREM1 Expression, Localization, and Interactions
FREM1 protein is expressed in a wide variety of human and animal cells and tissues. It functions as an important extracellular molecule either in developing embryos or in the pathogenesis of multiple abnormalities. FREM1 is primarily secreted by the mesenchymal cells underlying the basement membrane of visceral pleura and in epithelial and mesothelial cells [38][39]. The protein is localized in the dermal layer where it forms a stable complex with other Fras/FREM-related proteins called ternary complexes [2]. Although both dermal and epidermal cells express FREM1 [1][39], its expression is most prominent in dermal cells of the eyelids, head, and limbs of mice [3]. It is also expressed in the diaphragm [38], intestine, lung, kidney, and skin [39], as well as craniofacial structures, such as ears, forehead, midface, nose, teeth, and hair follicles in mice [3][40][41].
A previous study from our group showed that mRNA of FREM1 is expressed in various human tissues, including cervix, colon, esophagus, heart, kidney, lung, ovary, placenta, prostate, skeletal, small intestine, testes, thymus, thyroid, and trachea [15]. The expression of FREM1 was observed at a high level in cervical tissues, the small intestine, and kidneys. Immunohistochemistry staining of human ectocervical biopsy specimens from two different groups of women (HIV-1 negative new enrollees and HIV-1 resistant) of the Pumwani sex worker cohort demonstrated that FREM1 protein is expressed in the epithelial and sub-mucosal layer underneath the basement membrane (BM), particularly the upper lamina propria [15].
2.3. FREM1 and Immune Cell Infiltration
Extracellular matrix (ECM) proteins function as mediators of inflammation and infiltration of immune cells [42][43][44][45]. Because FREM1 is an extracellular matrix protein and widely expressed in the regions of epithelial–mesenchymal interaction [3], its expression in epithelial tissues may promote immune cell infiltration. Using immunohistochemistry (IHC) and immunofluorescence (IF) assays, a study demonstrated that elevated expression of FREM1 in normal breast/mammary epithelial tissues is positively correlated with the infiltration of several immune cells, such as CD4+, and CD8+T -cells, and M1 (inflammatory) macrophages [46]. In addition, analysis of breast cancer (BC) epithelial tissues showed that high FREM1-expressed BC tissues harbor abundant CD4+T memory cells (resting and activated), CD8+T-cells, M1 macrophages, resting dendritic cells, gamma-delta T cells (γδT), B-cells (naive and memory), plasma cells, and resting mast cells. Contrastingly, low FREM1-expressed BC tissues tended to have a higher proportion of M2 (anti-inflammatory) macrophages, neutrophils, and resting NK (natural killer) cells [46]. It has also been shown that the RGD motif of ECM binds with integrins and promotes migration of effector T cells into the interstitial inflamed tissues [47]. Thus, the binding of FREM1 with integrins may function as a critical regulator in infiltrating immune cells, especially in cervicovaginal tissues. Although the direct role of FREM1 in infiltrating immune cells in cervical tissues is yet to be examined, the expression of FREM1 in the cervix of humans [15] supports the notion that FREM1 may function as an inducer of immune cell infiltration, including HIV-1 target cells, facilitating vaginal HIV-1 infection.
2.4. FREM1 Polymorphism and HIV-1 Resistance
Pumwani sex worker cohort (PSWC) was established in the heart of Pumwani slum, Nairobi, Kenya in 1985. It is an open prospective cohort study of the immunobiology and epidemiology of sexually transmitted infections (STIs), and the patients enrolled in the cohort have been followed biannually since the cohort’s establishment [11][12][13][14]. This cohort is not only involved in the research, but also provides services related to STIs and HIV-1 prevention and care, such as consultation, provision of free condoms, and treatment of other infections. Between 1985 and 2012, more than 5000 sex workers have been enrolled in the cohort. A sub-group of women in PSWC remained HIV-1 uninfected despite the frequent exposure to high-risk HIV-1 infected sexual partners [14]. These women demonstrated a significantly high-level expression of various potentially HIV-1 inhibitory molecules, including RANTES (regulated on activation, normal T-cell expressed and secreted) [48], serpins, elafin, and many other factors [48][49][50] at their genital mucosa. Multiple immunological [51][52][53][54], genetic [7][8][9][10][55], and proteomics [49][50] factors are associated with this phenotype.
Our group has conducted a genome-wide analysis of SNP (single nucleotide polymorphism) among women of PSWC to identify the genetic polymorphism associated with the HIV-1 resistant phenotype [15]. The study showed that the minor allele of FREM1 SNP rs1552896 is enriched in HIV-1 resistant women and the major allele of the SNP rs1552896 is associated with HIV-1 susceptible women [15]. Furthermore, the frequency of the minor allele of rs1552896 was also found to be higher in HIV-1 uninfected women in comparison to HIV-1 infected individuals in a mother–child HIV-1 transmission (MCHT) cohort [15]. Thus, the minor allele of FREM1 SNP rs1552896 is associated with the HIV-1 resistant phenotype.
The SNP, rs1552896, is located at the intron 11 (57 bp) close to the 3′ end of the exon 11 of the FREM1 gene. The association of its minor allele with resistance to HIV-1 infection may be due to its influence in alternative splicing of FREM1, and/or it may be a marker for polymorphisms in the coding region that affects the structure and function of FREM1. To clarify this, we conducted full gene sequencing and a pilot RNA-Seq analysis in the women of the Pumwani sex worker cohort. Further genetic analysis identified a novel microsatellite (Tuff-ST) in LD (linkage disequilibrium) with rs1552896, and the allele SThomo of Tuff-ST microsatellite is strongly associated with HIV-1 resistant women and has a strong likelihood of modifying FREM1 transcript alternative splicing (Figure 2A) (Luo et al., unpublished data). Pilot RNA-Seq analysis supported this possibility and showed that the FREM1 isoform coding for TILRR was detected in most women with rs1552896-Tuff-ST wild type. However, this isoform is either absent or expressed at a very low level in women with the protective rs1552896-Tuff-ST (SThomo) genotype (p = 0.0067) (Figure 2B) (Luo et al., unpublished data). Based on these supporting data, we hypothesize that FREM1 plays an important role in the vaginal transmission of HIV-1 through the regulation of genes involved in immune activation, inflammatory responses, mucosal integrity, and cell migration. The association of the rs1552896-Tuff-ST (SThomo) genotype with resistance to HIV-1 infection may be partly due to an impaired IL-1R1/TLR signal transduction resulting from the abolition/reduced TILRR isoform expression.
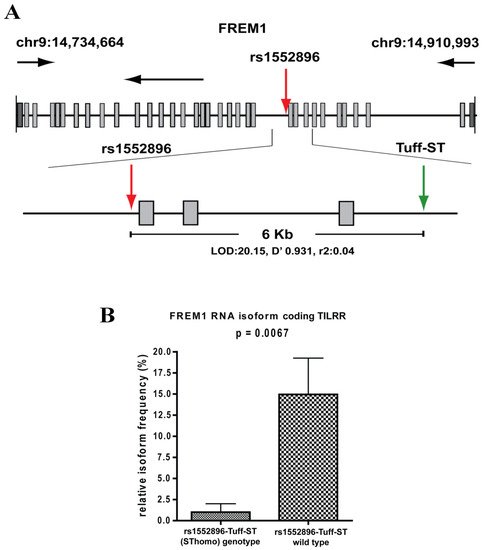
Figure 2. Full gene sequencing of FREM1 and RNA-Seq analysis of PBMCs. (A) Full gene sequencing of FREM1 gene of 70 women in the Pumwani sex worker cohort identified a novel microsatellite Tuff-ST that is in linkage disequilibrium with the SNP rs1552896 (LOD 20.15, D′ 0.931, r2: 0.04). Further Sanger sequencing and genotype of 1090 women for this microsatellite showed that rs1552896-Tuff-ST (SThomo) genotype is significantly associated with resistant women (p = 0.0002). (B) RNA-Seq analysis of PBMCs of Pumwani sex workers with different rs1552896-Tuff-ST genotypes. Women with the protective rs1552896-Tuff-ST (SThomo) genotype do not express or express a very low amount of FREM1 RNA isoform encoding TILRR in comparison with women with the rs1552896-Tuff-ST wild type (p = 0.0067) (Luo et al., unpublished data).
2.5. Monoclonal Antibodies to Study FREM1 and Its Splice Variant TILRR
Antibodies are important tools to study the function of proteins and have been used as therapeutics for many diseases [56]. To study the function of FREM1 and its role in HIV-1 infection, our group developed anti-FREM1 antibodies [28]. To develop anti-FREM1 monoclonal antibodies, we expressed two recombinant proteins of FREM1, rspD (376 amino acids, 56.8 kDa) and rspF (255 amino acids, 29.5 kDa). The two recombinant proteins overlap the C-terminal region of full-length FREM1 isoform 1 and the majority portion of FREM1 isoform 2 (TILRR). Structurally, rspD protein contains a small portion of the CSPG9 domain as well as CSPG10, CSPG11, CSPG12, and Calx-β domains of FREM1, whereas rspF only contains C-type Lectin domain (CTLD) (Figure 1C). Our group developed 17 anti-FREM1 mouse monoclonal antibodies (mAbs) and mapped their epitopes [28]. These anti-FREM1 mAbs target major and minor epitopes of multiple domains of FREM1 and its splice variants. These in-house developed anti-FREM1 mAbs and recombinant FREM1 proteins are tools to study the role of FREM1 and its variants in inflammation responses and vaginal HIV-1/SIV (simian immunodeficiency virus) acquisition. We are using this panel of mAbs to study the expression and function of FREM1 and TILRR.
This entry is adapted from the peer-reviewed paper 10.3390/ijms22157825
References
- Short, K.; Wiradjaja, F.; Smyth, I. Let‘s stick together: The role of the Fras1 and Frem proteins in epidermal adhesion. IUBMB Life 2007, 59, 427–435.
- Kiyozumi, D.; Sugimoto, N.; Sekiguchi, K. Breakdown of the reciprocal stabilization of QBRICK/Frem1, Fras1, and Frem2 at the basement membrane provokes Fraser syndrome-like defects. Proc. Natl. Acad. Sci. USA 2006, 103, 11981–11986.
- Smyth, I.; Du, X.; Taylor, M.S.; Justice, M.J.; Beutler, B.; Jackson, I.J. The extracellular matrix gene Frem1 is essential for the normal adhesion of the embryonic epidermis. Proc. Natl. Acad. Sci. USA 2004, 101, 13560–13565.
- Zhang, X.; Shephard, F.; Kim, H.B.; Palmer, I.R.; McHarg, S.; Fowler, G.J.; O’Neill, L.A.; Kiss-Toth, E.; Qwarnstrom, E.E. TILRR, a novel IL-1RI co-receptor, potentiates MyD88 recruitment to control Ras-dependent amplification of NF-kappaB. J. Biol. Chem. 2010, 285, 7222–7232.
- Chacon-Camacho, O.F.; Zenker, M.; Schanze, D.; Ledesma-Gil, J.; Zenteno, J.C. Novel FREM1 mutations in a patient with MOTA syndrome: Clinical findings, mutation update and review of FREM1-related disorders literature. Eur. J. Med. Genet. 2017, 60, 190–194.
- Zhang, X.; Pino, G.M.; Shephard, F.; Kiss-Toth, E.; Qwarnstrom, E.E. Distinct control of MyD88 adapter-dependent and Akt kinase-regulated responses by the interleukin (IL)-1RI co-receptor, TILRR. J. Biol. Chem. 2012, 287, 12348–12352.
- Lacap, P.A.; Huntington, J.D.; Luo, M.; Nagelkerke, N.J.; Bielawny, T.; Kimani, J.; Wachihi, C.; Ngugi, E.N.; Plummer, F.A. Associations of human leukocyte antigen DRB with resistance or susceptibility to HIV-1 infection in the Pumwani Sex Worker Cohort. AIDS 2008, 22, 1029–1038.
- Price, H.; Lacap, P.; Tuff, J.; Wachihi, C.; Kimani, J.; Ball, T.B.; Luo, M.; Plummer, F.A. A TRIM5alpha exon 2 polymorphism is associated with protection from HIV-1 infection in the Pumwani sex worker cohort. AIDS 2010, 24, 1813–1821.
- Hardie, R.A.; Knight, E.; Bruneau, B.; Semeniuk, C.; Gill, K.; Nagelkerke, N.; Kimani, J.; Wachihi, C.; Ngugi, E.; Luo, M.; et al. A common human leucocyte antigen-DP genotype is associated with resistance to HIV-1 infection in Kenyan sex workers. AIDS 2008, 22, 2038–2042.
- Hardie, R.A.; Luo, M.; Bruneau, B.; Knight, E.; Nagelkerke, N.J.; Kimani, J.; Wachihi, C.; Ngugi, E.N.; Plummer, F.A. Human leukocyte antigen-DQ alleles and haplotypes and their associations with resistance and susceptibility to HIV-1 infection. AIDS 2008, 22, 807–816.
- Simonsen, J.N.; Plummer, F.A.; Ngugi, E.N.; Black, C.; Kreiss, J.K.; Gakinya, M.N.; Waiyaki, P.; D’Costa, L.J.; Ndinya-Achola, J.O.; Piot, P.; et al. HIV infection among lower socioeconomic strata prostitutes in Nairobi. AIDS 1990, 4, 139–144.
- Kreiss, J.K.; Koech, D.; Plummer, F.A.; Holmes, K.K.; Lightfoote, M.; Piot, P.; Ronald, A.R.; Ndinya-Achola, J.O.; D’Costa, L.J.; Roberts, P.; et al. AIDS virus infection in Nairobi prostitutes. Spread of the epidemic to East Africa. N. Engl. J. Med. 1986, 314, 414–418.
- Plummer, F.A.; Simonsen, J.N.; Cameron, D.W.; Ndinya-Achola, J.O.; Kreiss, J.K.; Gakinya, M.N.; Waiyaki, P.; Cheang, M.; Piot, P.; Ronald, A.R.; et al. Cofactors in male-female sexual transmission of human immunodeficiency virus type 1. J. Infect. Dis. 1991, 163, 233–239.
- Fowke, K.R.; Nagelkerke, N.J.; Kimani, J.; Simonsen, J.N.; Anzala, A.O.; Bwayo, J.J.; MacDonald, K.S.; Ngugi, E.N.; Plummer, F.A. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet 1996, 348, 1347–1351.
- Luo, M.; Sainsbury, J.; Tuff, J.; Lacap, P.A.; Yuan, X.Y.; Hirbod, T.; Kimani, J.; Wachihi, C.; Ramdahin, S.; Bielawny, T.; et al. A genetic polymorphism of FREM1 is associated with resistance against HIV infection in the Pumwani sex worker cohort. J. Virol. 2012, 86, 11899–11905.
- Kashem, M.A.; Li, H.; Toledo, N.P.; Omange, R.W.; Liang, B.; Liu, L.R.; Li, L.; Yang, X.; Yuan, X.-Y.; Kindrachuk, J.; et al. Toll-like Interleukin 1 Receptor Regulator Is an Important Modulator of Inflammation Responsive Genes. Front. Immunol. 2019, 10, 1–16.
- Fichorova, R.N.; Tucker, L.D.; Anderson, D.J. The molecular basis of nonoxynol-9-induced vaginal inflammation and its possible relevance to human immunodeficiency virus type 1 transmission. J. Infect. Dis. 2001, 184, 418–428.
- Dabee, S.; Barnabas, S.L.; Lennard, K.S.; Jaumdally, S.Z.; Gamieldien, H.; Balle, C.; Happel, A.U.; Murugan, B.D.; Williamson, A.L.; Mkhize, N.; et al. Defining characteristics of genital health in South African adolescent girls and young women at high risk for HIV infection. PLoS ONE 2019, 14, e0213975.
- Cohen, C.R.; Moscicki, A.B.; Scott, M.E.; Ma, Y.; Shiboski, S.; Bukusi, E.; Daud, I.; Rebbapragada, A.; Brown, J.; Kaul, R. Increased levels of immune activation in the genital tract of healthy young women from sub-Saharan Africa. AIDS 2010, 24, 2069–2074.
- Kaul, R.; Prodger, J.; Joag, V.; Shannon, B.; Yegorov, S.; Galiwango, R.; McKinnon, L. Inflammation and HIV Transmission in Sub-Saharan Africa. Curr. HIV/AIDS Rep. 2015, 12, 216–222.
- Pan, X.; Baldauf, H.M.; Keppler, O.T.; Fackler, O.T. Restrictions to HIV-1 replication in resting CD4+ T lymphocytes. Cell Res. 2013, 23, 876–885.
- Biancotto, A.; Iglehart, S.J.; Vanpouille, C.; Condack, C.E.; Lisco, A.; Ruecker, E.; Hirsch, I.; Margolis, L.B.; Grivel, J.C. HIV-1 induced activation of CD4+ T cells creates new targets for HIV-1 infection in human lymphoid tissue ex vivo. Blood 2008, 111, 699–704.
- Zhang, Z.Q.; Wietgrefe, S.W.; Li, Q.; Shore, M.D.; Duan, L.; Reilly, C.; Lifson, J.D.; Haase, A.T. Roles of substrate availability and infection of resting and activated CD4+ T cells in transmission and acute simian immunodeficiency virus infection. Proc. Natl. Acad. Sci. USA 2004, 101, 5640–5645.
- Passmore, J.A.; Jaspan, H.B.; Masson, L. Genital inflammation, immune activation and risk of sexual HIV acquisition. Curr. Opin. HIV AIDS 2016, 11, 156–162.
- Masson, L.; Passmore, J.A.; Liebenberg, L.J.; Werner, L.; Baxter, C.; Arnold, K.B.; Williamson, C.; Little, F.; Mansoor, L.E.; Naranbhai, V.; et al. Genital inflammation and the risk of HIV acquisition in women. Clin. Infect. Dis. 2015, 61, 260–269.
- Kiyozumi, D.; Osada, A.; Sugimoto, N.; Weber, C.N.; Ono, Y.; Imai, T.; Okada, A.; Sekiguchi, K. Identification of a novel cell-adhesive protein spatiotemporally expressed in the basement membrane of mouse developing hair follicle. Exp. Cell Res. 2005, 306, 9–23.
- Staub, E.; Hinzmann, B.; Rosenthal, A. A novel repeat in the melanoma-associated chondroitin sulfate proteoglycan defines a new protein family. FEBS Lett. 2002, 527, 114–118.
- Yuan, X.Y.; Liu, L.R.; Krawchenko, A.; Sainsbury, J.; Zhao, L.; Plummer, F.; Yang, X.; Luo, M. Development of monoclonal antibodies to interrogate functional domains and isoforms of FREM1 protein. Monoclon. Antib. Immunodiagn. Immunother. 2014, 33, 129–140.
- Nishiyama, A.; Dahlin, K.J.; Prince, J.T.; Johnstone, S.R.; Stallcup, W.B. The primary structure of NG2, a novel membrane-spanning proteoglycan. J. Cell Biol. 1991, 114, 359–371.
- Burg, M.A.; Tillet, E.; Timpl, R.; Stallcup, W.B. Binding of the NG2 proteoglycan to type VI collagen and other extracellular matrix molecules. J. Biol. Chem. 1996, 271, 26110–26116.
- Tillet, E.; Gential, B.; Garrone, R.; Stallcup, W.B. NG2 proteoglycan mediates beta1 integrin-independent cell adhesion and spreading on collagen VI. J. Cell Biochem. 2002, 86, 726–736.
- Nishiyama, A.; Stallcup, W.B. Expression of NG2 proteoglycan causes retention of type VI collagen on the cell surface. Mol. Biol. Cell 1993, 4, 1097–1108.
- Goretzki, L.; Burg, M.A.; Grako, K.A.; Stallcup, W.B. High-affinity binding of basic fibroblast growth factor and platelet-derived growth factor-AA to the core protein of the NG2 proteoglycan. J. Biol. Chem. 1999, 274, 16831–16837.
- Nathanson, J.; Swarr, D.T.; Singer, A.; Liu, M.; Chinn, A.; Jones, W.; Hurst, J.; Khalek, N.; Zackai, E.; Slavotinek, A. Novel FREM1 mutations expand the phenotypic spectrum associated with Manitoba-oculo-tricho-anal (MOTA) syndrome and bifid nose renal agenesis anorectal malformations (BNAR) syndrome. Am. J. Med. Genet. A 2013, 161A, 473–478.
- Schwarz, E.M.; Benzer, S. Calx, a Na-Ca exchanger gene of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1997, 94, 10249–10254.
- Thierry-Mieg, D.; Thierry-Mieg, J. AceView: A comprehensive cDNA-supported gene and transcripts annotation. Genome. Biol. 2006, 7, 11–14.
- NCBI. The AceView Genes. Available online: https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/ (accessed on 20 April 2020).
- Beck, T.F.; Veenma, D.; Shchelochkov, O.A.; Yu, Z.; Kim, B.J.; Zaveri, H.P.; van Bever, Y.; Choi, S.; Douben, H.; Bertin, T.K.; et al. Deficiency of FRAS1-related extracellular matrix 1 (FREM1) causes congenital diaphragmatic hernia in humans and mice. Hum. Mol. Genet. 2013, 22, 1026–1038.
- Petrou, P.; Chiotaki, R.; Dalezios, Y.; Chalepakis, G. Overlapping and divergent localization of Frem1 and Fras1 and its functional implications during mouse embryonic development. Exp. Cell Res. 2007, 313, 910–920.
- Vissers, L.E.; Cox, T.C.; Maga, A.M.; Short, K.M.; Wiradjaja, F.; Janssen, I.M.; Jehee, F.; Bertola, D.; Liu, J.; Yagnik, G.; et al. Heterozygous mutations of FREM1 are associated with an increased risk of isolated metopic craniosynostosis in humans and mice. PLoS Genet. 2011, 7, e1002278.
- Alazami, A.M.; Shaheen, R.; Alzahrani, F.; Snape, K.; Saggar, A.; Brinkmann, B.; Bavi, P.; Al-Gazali, L.I.; Alkuraya, F.S. FREM1 mutations cause bifid nose, renal agenesis, and anorectal malformations syndrome. Am. J. Hum. Genet. 2009, 85, 414–418.
- Arroyo, A.G.; Iruela-Arispe, M.L. Extracellular matrix, inflammation, and the angiogenic response. Cardiovasc. Res. 2010, 86, 226–235.
- Adair-Kirk, T.L.; Senior, R.M. Fragments of extracellular matrix as mediators of inflammation. Int. J. Biochem. Cell Biol. 2008, 40, 1101–1110.
- Gaudet, A.D.; Popovich, P.G. Extracellular matrix regulation of inflammation in the healthy and injured spinal cord. Exp. Neurol. 2014, 258, 24–34.
- Gopal, S. Syndecans in Inflammation at a Glance. Front. Immunol. 2020, 11, 227.
- Li, H.N.; Li, X.R.; Lv, Z.T.; Cai, M.M.; Wang, G.; Yang, Z.F. Elevated expression of FREM1 in breast cancer indicates favorable prognosis and high-level immune infiltration status. Cancer Med. 2020.
- Kim, C.H. Crawling of effector T cells on extracellular matrix: Role of integrins in interstitial migration in inflamed tissues. Cell Mol. Immunol. 2014, 11, 1–4.
- Iqbal, S.M.; Ball, T.B.; Kimani, J.; Kiama, P.; Thottingal, P.; Embree, J.E.; Fowke, K.R.; Plummer, F.A. Elevated T cell counts and RANTES expression in the genital mucosa of HIV-1-resistant Kenyan commercial sex workers. J. Infect. Dis. 2005, 192, 728–738.
- Burgener, A.; Boutilier, J.; Wachihi, C.; Kimani, J.; Carpenter, M.; Westmacott, G.; Cheng, K.; Ball, T.B.; Plummer, F. Identification of differentially expressed proteins in the cervical mucosa of HIV-1-resistant sex workers. J. Proteome. Res. 2008, 7, 4446–4454.
- Iqbal, S.M.; Ball, T.B.; Levinson, P.; Maranan, L.; Jaoko, W.; Wachihi, C.; Pak, B.J.; Podust, V.N.; Broliden, K.; Hirbod, T.; et al. Elevated elafin/trappin-2 in the female genital tract is associated with protection against HIV acquisition. AIDS 2009, 23, 1669–1677.
- Alimonti, J.B.; Kimani, J.; Matu, L.; Wachihi, C.; Kaul, R.; Plummer, F.A.; Fowke, K.R. Characterization of CD8 T-cell responses in HIV-1-exposed seronegative commercial sex workers from Nairobi, Kenya. Immunol. Cell Biol. 2006, 84, 482–485.
- Alimonti, J.B.; Koesters, S.A.; Kimani, J.; Matu, L.; Wachihi, C.; Plummer, F.A.; Fowke, K.R. CD4+ T cell responses in HIV-exposed seronegative women are qualitatively distinct from those in HIV-infected women. J. Infect. Dis. 2005, 191, 20–24.
- Luo, M.; Daniuk, C.A.; Diallo, T.O.; Capina, R.E.; Kimani, J.; Wachihi, C.; Kimani, M.; Bielawny, T.; Peterson, T.; Mendoza, M.G.; et al. For protection from HIV-1 infection, more might not be better: A systematic analysis of HIV Gag epitopes of two alleles associated with different outcomes of HIV-1 infection. J. Virol. 2012, 86, 1166–1180.
- Rowland-Jones, S.L.; Dong, T.; Fowke, K.R.; Kimani, J.; Krausa, P.; Newell, H.; Blanchard, T.; Ariyoshi, K.; Oyugi, J.; Ngugi, E.; et al. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J. Clin. Investig. 1998, 102, 1758–1765.
- Ball, T.B.; Ji, H.; Kimani, J.; McLaren, P.; Marlin, C.; Hill, A.V.; Plummer, F.A. Polymorphisms in IRF-1 associated with resistance to HIV-1 infection in highly exposed uninfected Kenyan sex workers. AIDS 2007, 21, 1091–1101.
- Salazar, G.; Zhang, N.; Fu, T.M.; An, Z. Antibody therapies for the prevention and treatment of viral infections. NPJ Vaccines 2017, 2, 19.
This entry is offline, you can click here to edit this entry!
