In the search for new therapeutic strategies to contrast SARS-CoV-2, we here studied the interaction of polydatin (PD) and resveratrol (RESV)—two natural stilbene polyphenols with manifold, well known biological activities—with Spike, the viral protein essential for virus entry into host cells, and ACE2, the angiotensin-converting enzyme present on the surface of multiple cell types (including respiratory epithelial cells) which is the main host receptor for Spike binding. Molecular Docking simulations evidenced that both compounds can bind Spike, ACE2 and the ACE2:Spike complex with good affinity, although the interaction of PD appears stronger than that of RESV on all the investigated targets. Preliminary biochemical assays revealed a significant inhibitory activity of the ACE2:Spike recognition with a dose-response effect only in the case of PD.
- SARS-CoV-2
- polydatin
- resveratrol
- molecular docking
- protein-binding
- ACE2:Spike binding-inhibition
1. Introduction
2. Molecular docking simulation.
Spike-protein pre-fusion conformation [50,76] is a trimer constituted of two subunits, S1 and S2, which are cleaved following receptor binding [77]. S1 Receptor Binding Domains (RBDs) host the binding motifs (RBMs) able to recognize ACE2. The high RBD flexibility allows the Spike to sample open or closed conformations, in which RBMs are respectively exposed or hidden inside the protomers interface [77–81].
Therefore, the binding to SARS-CoV-2 Spike structure with one RBD in an open conformation (PDB ID: 6VSB [50]) has been investigated by molecular docking simulations. The pockets on the RBD surface appear able to accommodate both PD and RESV ligands (Figure 2). A slightly lower affinity for the RBD domain was found for RESV (−6.5 kcal/mol for the best docking pose, Table 1) with respect to PD (top-ranked pose −6.9 kcal/mol).
Table 1. Docking scores (i.e., the approximate binding energy estimated by the docking scoring function, kcal/mol units) for PD and RESV top-ranked docked poses.
|
|
PD |
RESV |
|
Spike RBD |
−6.9 |
−6.5 |
|
ACE2 |
−8.4 |
−6.9 |
|
S:ACE2 region I |
−8.1 |
−7.6 |
|
S:ACE2 region II |
−6.9 |
−6.5 |
ACE2 homodimer (PDB ID: 6M18, [51]) is constituted by an N-terminal protease domain, involved in the interaction with SARS-CoV-2 Spike RBD [51], and a C-terminal collectrin-like domain, comprising the ACE2 transmembrane helix. Molecular docking simulations were performed near ACE2 protease domain α1 and α2 helices, thus far from the catalytic site related to ACE2 physiological function. Despite its size, PD can be easily accommodated in a deep groove behind the two helices (Figure 3a,b) producing a high docking score (−8.4 kcal/mol, Table 1). RESV can also bind to the pockets near α1 and α2 helices, but with a ~1 kcal/mol lower score with respect to PD (−6.9 kcal/mol, Table 1) (Figure 3c,d).
Both RESV and PD showed a good estimated binding affinity in the groove between the Spike RBD and ACE2. Due to the high spatial extent of the Spike:ACE2 interface, docking simulations were performed in two distinct regions (named regions I and II). In the first interfacial region investigated (region I), the most stable RESV binding mode involves recognition of one of the interfacial pockets through polar and hydrophobic contacts (Figure 4c,d). PD binds in the same pocket as RESV (Figure 4a,b) but interacting with more surrounding residues, thus showing a higher docking score (−8.1 kcal/mol, Table 1), compared to that of RESV (−7.6 kcal/mol).
In the second region (region II) investigated at the interface between viral Spike RBD and human ACE2, the top-ranked RESV conformer fits into a large ACE2 cavity far from the interaction sites between the two protein domains. On the contrary, in the best PD binding mode, the ligand binds onto a pocket located at one end of the ACE2:Spike complex interface with a docking score 0.4 kcal/mol lower than RESV (−6.9 kcal/mol, Table 1). It is interesting to note the fundamental role played by the glucosidic moiety which interacts with the residues of the main chains that are involved in the formation of the ACE2:Spike dimer. Weakening of these interactions could, in principle, contribute to the dissociation of the dimeric complex.
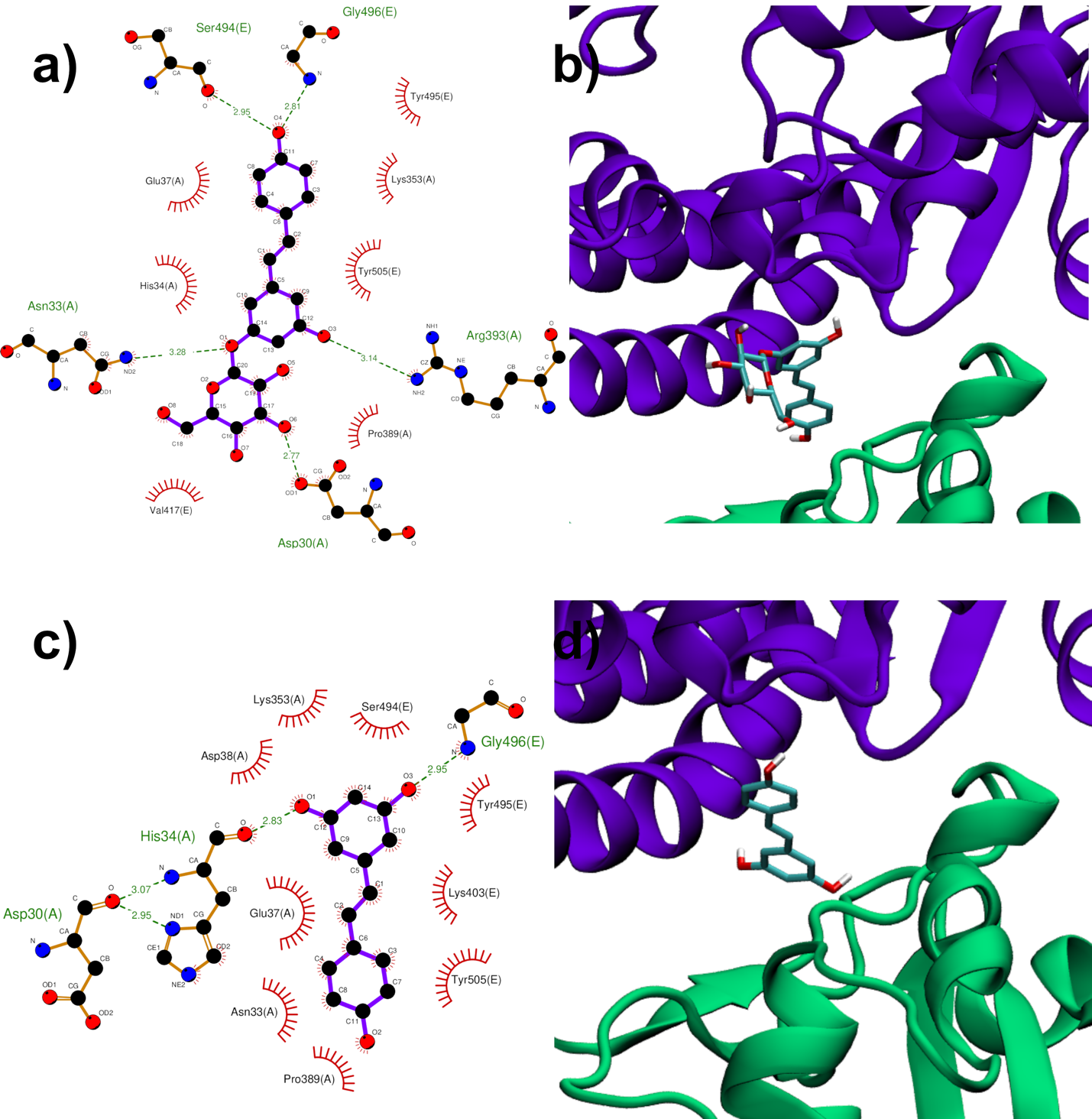
Figure 4. PD (a,b) and RESV (c,d) top-ranked poses docked to Spike:ACE2 interface region I. Two-dimensional interaction maps (a–c): C, N and O atoms are reported in black, blue and red, respectively. Hydrogen bonds are depicted as green dashed lines, while hydrophobic interactions as red cogwheels. Hydrogen atoms are not depicted for ease of illustration. The names of proteins’ residues involved in interactions with the ligand are reported. Three-dimensional representations of PD and RESV docked poses (b–d): proteins’ backbone is represented as a cartoon, while the ligand as sticks.
Results from docking simulations on the already assembled Spike:ACE2 complex reveal the potential capability of PD, but also RESV, to insert themselves into the extended adduct interface. This leads to the hypothesis of a ligand-induced dissociation or weakening effect, through allosteric (such as for region I) or direct (region II) interference.
3. To establish if RESV and PD can experimentally interfere with the binding of the Spike protein with ACE2 receptor, as suggested by the molecular docking simulations, binding inhibition assays were performed.
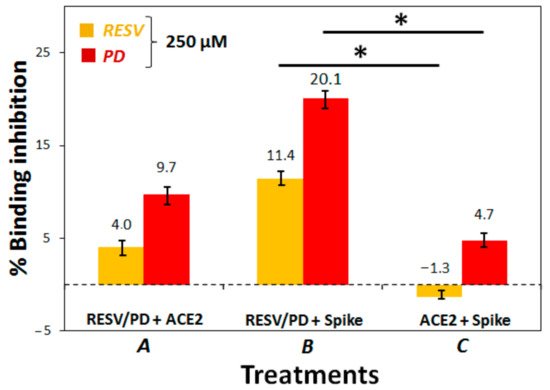
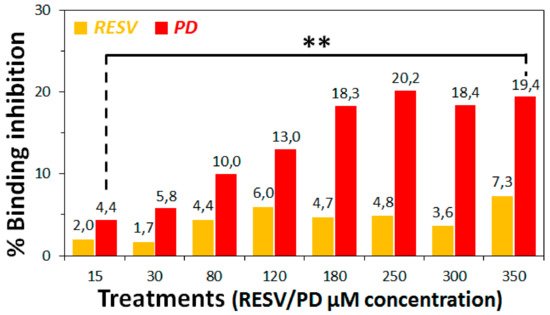
4. Conclusions
This entry is adapted from the peer-reviewed paper 10.3390/biom11071048
