Exploitation of Heterosis in Pearl Millet: A Review
by Rakesh K. Srivastava *, Srikanth Bollam, Vijayalakshmi Pujarula, Madhu Pusuluri, Ram B. Singh, Gopi Potupureddi and Rajeev Gupta *
International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Hyderabad TS 502324, India
*
Authors to whom correspondence should be addressed.
Plants 2020, 9(7), 807; https://doi.org/10.3390/plants9070807
Received: 6 March 2020 / Revised: 23 June 2020 / Accepted: 23 June 2020 / Published: 27 June 2020
Abstract
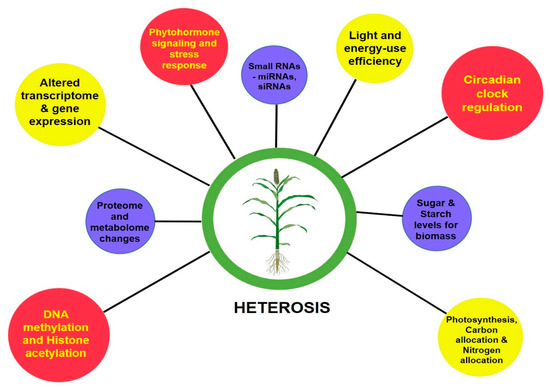
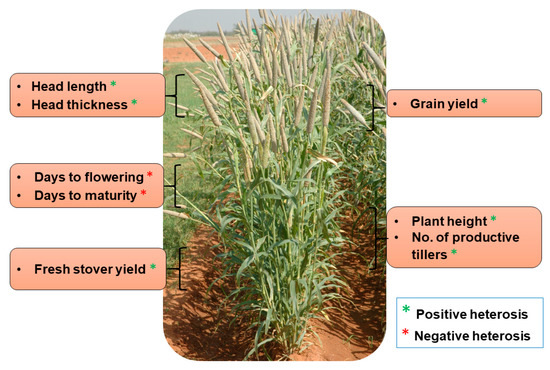
The phenomenon of heterosis has fascinated plant breeders ever since it was first described by Charles Darwin in 1876 in the vegetable kingdom and later elaborated by George H Shull and Edward M East in maize during 1908. Heterosis is the phenotypic and functional superiority manifested in the F1 crosses over the parents. Various classical complementation mechanisms gave way to the study of the underlying potential cellular and molecular mechanisms responsible for heterosis. In cereals, such as maize, heterosis has been exploited very well, with the development of many single-cross hybrids that revolutionized the yield and productivity enhancements. Pearl millet (Pennisetum glaucum (L.) R. Br.) is one of the important cereal crops with nutritious grains and lower water and energy footprints in addition to the capability of growing in some of the harshest and most marginal environments of the world. In this highly cross-pollinating crop, heterosis was exploited by the development of a commercially viable cytoplasmic male-sterility (CMS) system involving a three-lines breeding system (A-, B- and R-lines). The first set of male-sterile lines, i.e., Tift 23A and Tift18A, were developed in the early 1960s in Tifton, Georgia, USA. These provided a breakthrough in the development of hybrids worldwide, e.g., Tift 23A was extensively used by Punjab Agricultural University (PAU), Ludhiana, India, for the development of the first single-cross pearl millet hybrid, named Hybrid Bajra 1 (HB 1), in 1965. Over the past five decades, the pearl millet community has shown tremendous improvement in terms of cytoplasmic and nuclear diversification of the hybrid parental lines, which led to a progressive increase in the yield and adaptability of the hybrids that were developed, resulting in significant genetic gains. Lately, the whole genome sequencing of Tift 23D2B1 and re-sequencing of circa 1000 genomes by a consortium led by the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) has been a significant milestone in the development of cutting-edge genetic and genomic resources in pearl millet. Recently, the application of genomics and molecular technologies has provided better insights into genetic architecture and patterns of heterotic gene pools. Development of whole-genome prediction models incorporating heterotic gene pool models, mapped traits and markers have the potential to take heterosis breeding to a new level in pearl millet. This review discusses advances and prospects in various fronts of heterosis for pearl millet.
Keywords: heterosis; hybrid vigor; pearl millet; genome sequence; heterotic gene pools; genomic selection