Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Plant Sciences
The Retinoblastoma protein (pRb) is a key cell cycle regulator conserved in a wide variety of organisms. Experimental analysis of pRb’s functions in animals and plants has revealed that this protein participates in cell proliferation and differentiation processes. In addition, pRb in animals and its orthologs in plants (RBR), are part of highly conserved protein complexes which suggest the possibility that analogies exist not only between functions carried out by pRb orthologs themselves, but also in the structure and roles of the protein networks where these proteins are involved.
- Retinoblastoma protein
- cell cycle
- Arabidopsis
- humans
- differentiation
- Epigenetic
- DNA damage
- Protein structure
- Retinoblastoma related
Structure of the Retinoblastoma Protein
In humans, Retinoblastoma susceptibility gene is a member of a small gene family that includes RB1 (p105/pRb), RBl1 (p107/pRBL1), and RBl2 (p130/pRBL2), whose protein structure are very similar, and that share some overlapping functions [1][2][3].
The human Retinoblastoma protein (pRb) consists of 928 amino acids and includes three distinctive domains: the N-terminal structural domain (RbN), the so-called “pocket” (RbP) domain, the C-terminal domain (RbC), and the non-structured regions between them (Figure 1A). The pocket domain includes two highly conserved subdomains (A and B) called cyclin folds, which are formed by two structural nuclei, each conformed by three helix bundles with two additional helices packing at the sides in each one. These subdomains are required to mediate interactions with other proteins like several oncoproteins and transcription factors (TFs) [4][5][6][7]. According to current interaction databases 322 proteins interact with human pRb, the E2F TFs being the best characterized ones (Figure 1A) [8][9]. Many of the pRb-interacting proteins contain the motif ‘LxCxE’ (Leu-X-Cys-X-Glu where X stands for any amino acid), essential to bind with the Pocket B subdomain of pRb (Figure 1A).
In total, human pRb has 14 phosphorylation sites [7][10], two acetylation sites [11][12], six methylation sites [13][14][15], and it can also be modified through ubiquitination or sumoylation [16][17][18] (Figure 1B). Studies on the structure of pRb have revealed that phosphorylation changes pRb structure and promotes new interactions with other proteins.
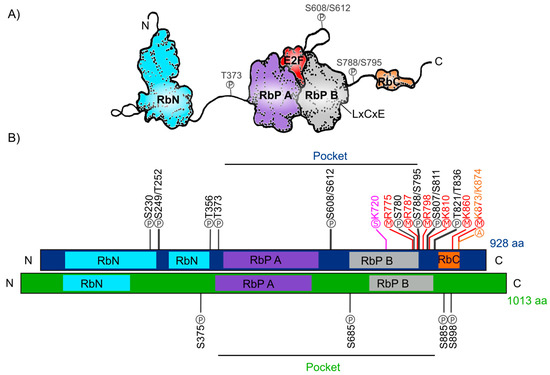
Figure 1. Retinoblastoma protein structure. (A) Representation of the human Rb protein structure with the domains RbN (blue), Pocket (RbP), with the RbP A (purple) and B (grey) subdomains interacting with an E2F TF (red), the RbC domain (orange) and the inter-domains (black lines) are also shown. The “P” inside a circle represents three examples of phosphorylation sites that change the structure of the protein. The position of the LxCxE cleft that allows Retinoblastoma protein (pRb) to interact with different proteins is also shown. (B) Comparison of the domains of human Rb protein (blue foreground) and Arabidopsis RBR protein (green foreground) and their reported post-translational modifications. Phosphorylation sites (P) are shown in black, methylation sites (M) in red, acetylation sites (A) in orange, and sumoylation sites (S) in pink.
The RB ortholog in plants was identified approximately a decade after the animal gene was discovered: RB1 gene was first described in humans between 1986 and 1989 [19][20][21] and the first plant orthologous RB1 cDNA (RBR), was first identified and cloned from maize in 1994 (ZmRBR) [22][23][24], then in tobacco (NtRBR) [25] and then in Arabidopsis (AtRBR) [26]. Afterwards, there have been numerous reports characterizing RBR orthologs in different plant species. Intriguingly, monocotyledons seem to have various RBR paralogs while dicotyledons have only one [27]. As a dicot plant, Arabidopsis carries a single copy of the RBR gene displaying ≈35% sequence similarity within the pocket domain with respect to the human pRb [28][29][30]. The Arabidopsis RBR (AtRBR) protein contains 1013 amino acids with the same modular structure of human pRb with putative phosphorylation residues. The role of four of them, located in the protein’s inter-domains, have been experimentally tested (Figure 1B) [31][32][33].
Cell Cycle Control through pRb
pRb hypophosphorylated acts as a negative regulator of cell cycle progression through its interaction with the E2F proteins. The heterodimer keeps the chromatin in a closed conformation in the regulatory regions of E2F-regulated genes [34][35]. When the pRb/E2F interaction is disrupted by loss or reduction of pRb, a high rate of cell proliferation is observed, and this generally triggers cancer [36]. pRb phosphorylation by cyclin/CDKs (cyclin-dependent kinases) complexes changes the pRb protein structure and its interactions with other proteins, inducing the release of E2F. For instance, this occurs in human cells when cyclin type D or E (CYCD/E) associate with cyclin kinases 4 or 2 (CDK4/2), respectively [34][37][38]. When pRb phosphorylation is altered, for example by overexpression of CYCDs, or by a miss regulation of CDKs, or by a disruption of the LxCxE-binding function the cell cycle is altered [39][40][41].
In humans, pRBL1 and pRBL2 participate in the repression of genes when cells are in the G0 quiescent state, through a complex called DREAM (DP-Rb-E2F-MuvB), that coordinates the repression of genes during quiescence and also the periodic gene expression during the G1/S and G2/M transitions [42][43]. DREAM is a multimeric protein complex that in humans is composed of DP (DP1-DP3), pRBL1 or pRBL2, E2F proteins (E2F4 and E2F5), and the subcomplex MuvB (Multi vulval class B). The MuvB subcomplex acts as a repressor when it is part of the DREAM complex, and is composed of LIN (LIN9, LIN37, LIN52, LIN54) and RBBP4 proteins. When pRb is phosphorylated, the DREAM complex is disassembled and the MuvB subcomplex can associate with TFs such as B-Myb (Myb type) and Fox-M1 (Forkhead box protein M 1), to promote the regulation of gene expression and the transition from quiescence to proliferation [42][44][45].
At about the same time that the pRb ortholog in plants was discovered, homologs of other components of the animal’s cell cycle regulatory machinery were identified and characterized in corn, as well as in Arabidopsis and Medicago sativa (alfalfa) [46][47][48][49][50]. In these plants, it was shown that the phosphorylation of RBR by the CDKA/CYCD protein complex regulates cell cycle progression, as it occurs in humans [51][52].
The DREAM complex also appears to be conserved in plants [53][54]. In addition to the presence of AtRBR, E2Fs/DPs, a MuvB-like complex has also been found that contains ALY2/3 (orthologs of LIN9) and TCX5 that is part of the TSO1-like family members (orthologs of LIN54) [55][56]. Furthermore, MYB3R, a transcription factor of the Myb family, has a protein structure resembling the DNA binding domain of B-Myb, which in humans forms part of the MuvB complex. These plant’s MYB3Rs also participate in the regulation of the G2/M transition as follows: MYB3R3 associates with E2Fc and AtRBR to repress G2/M genes, while MYB3R4 associates with E2Fb and AtRBR to activate G2/M genes [57][58][59].
The Roles of pRb in Cell Differentiation
The formation of any organ relies on two different but interlinked cellular processes: cell proliferation and cell differentiation. Proliferation produces all the cells that later will acquire fates, specialized functions, and morphologies through differential gene expression during cell differentiation [60][61]. pRb has been widely studied in proliferation and, recently, its participation in many different animal differentiation processes in the eye, brain, peripheral nervous system, muscle, liver, placenta, lung, cerebellum, pituitary gland, and heart has been elucidated (Figure 2A) [62][63][64][65][66][67].
Similar to pRb in animals, RBR participates in differentiation processes in the root and shoot meristems, the vascular system, leaves, stomas, and trichomes tissues in plants (Figure 2A) [68][69][70][71]. In Arabidopsis, AtRBR functions as a negative regulator of primary root development; its downregulation leads to longer roots with larger meristems whereas its overexpression results in shorter roots with smaller meristems [72].
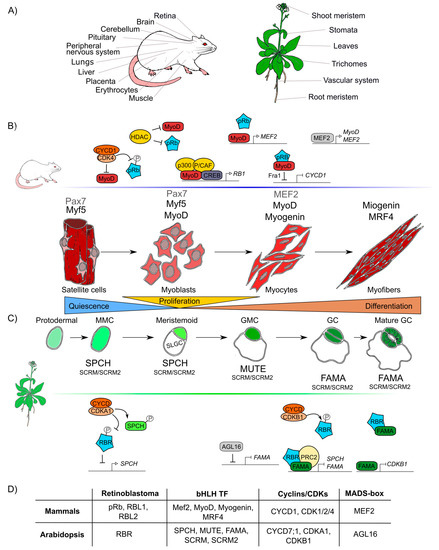
Figure 2. Retinoblastoma proteins are involved in cell differentiation in mammals and plants. (A) Mammals’ and plants’ organs whose differentiation depends on pRb/RBR. (B) Sequential steps of skeletal muscle differentiation in mammals. Shown are pRb interactions and bHLHs-family proteins (Mef2, MyoD, Myogenin, MRF4), involved in muscle quiescence maintenance, proliferation, and differentiation. (C) Sequential steps of guard cells differentiation in Arabidopsis. Shown are RBR interactions and bHLHs-family proteins (SPCH, MUTE, FAMA, SCRM, SCRM2) involved in quiescence maintenance, proliferation, and differentiation of guard cells. (D) Correlations between components involved in muscle and guard cell development in mammals and Arabidopsis, respectively.
It is difficult to compare the roles of pRb and RBR during differentiation due to lack of information and because the components involved in cell differentiation are more species-specific and less conserved than those involved in cell cycle regulation. However, taking as examples skeletal muscle differentiation in humans and stomatal guard cells differentiation in Arabidopsis, conserved factors that coordinate cell differentiation can be identified, namely Rb, MADS-box TFs, and TFs of the bHLH family, as well as the cyclins/CDKs protein complexes (Figure 2D) [73][74].
At post-embryonic stages, myocyte differentiation in mammals can be triggered in response to muscle damage or a specific growth-inducing stimulus. This last provokes massive proliferation of myoblasts through the hierarchical activation of several MRF (Myogenic Regulatory Factors): Myogenic Factor 5 (Myf5), myoblast determination protein 1 (MyoD), Myogenin (MyoG), and Myogenic Regulatory Factor 4B (MRF4), which are TFs that belong to the bHLH family [75][76][77]. In this process, the protein complex of MyoD with pRb, is thought to initiate differentiation, as it promotes cell cycle arrest [78][79][80][81]. Despite immunoprecipitation experiments that proved that pRb and MyoD can interact [82], later experiments using nuclear magnetic resonance allowed to determine that there is no direct protein-protein interaction between MyoD and pRb [83]. Still, MyoD does activate RB1 expression through its association with the cAMP response element-binding (CREB) TF and the coactivators p300 and P/CAF (Figure 2) [84][85].
Induction of muscle biogenesis also requires the regulation by cyclins-CDKs in association with pRb [86]. The stable repression of cyclin D1, required for cell cycle arrest during differentiation, is regulated by the joint action of MyoD and pRb through the regulation of the upstream intermediary gene Fra-1 (antigen related to FOS 1) (Figure 2B) [87][88]. Antagonistically, cyclin D1 inhibits the activity of MyoD: overexpression of cyclin D1 promotes nuclear accumulation of CDK4, that binds MyoD, preventing its interaction with DNA, and inhibiting the CDK4-dependent phosphorylation of pRb (Figure 2B) [89][90][91]. Additionally, when HDAC1 interacts with pRb, the MyoD protein can bind its target DNA regulatory sites [92][93]. In summary, there are two different protein complexes formed during different stages of muscle development: HDAC1/MyoD during proliferation and HDAC1/hypophosphorylated pRb during differentiation (Figure 2B) [92][93].
In plants, some epidermal cells undergo differentiation producing the two mature guard cells that form the stomata pores, structures that are conserved among land plants and allow them to regulate gas exchange and water loss [94][95]. Stomatal development is hierarchically regulated by the sequential activation of several TFs of the bHLH family: SPCH (SPEECHLESS), MUTE, and FAMA. These three bHLHs form heterodimers with either the bHLHs SCREAM (SCRM, also called ICE1) or SCRM2 [96][97][98], that belong to the same family of TFs that participate in muscle development in mammals (MRFs) (Figure 2D). These plant TFs orchestrate cell division events of protodermal cells, which give origin to guard cells (stomatogenesis). SPCH triggers the maturation of a protodermal cell into a meristemoid mother cell (MMC) and is also involved in the asymmetric cell division of the MMC, that results in one meristemoid cell and one larger sister cell (SLGC). The meristemoid cell exits stemness and engages in differentiation to become a guard meristemoid cell (GMC). MUTE must be expressed at this stage, to direct further differentiation of a GMC, and to ensure that this cell undergoes a single symmetric division. In addition MUTE regulates the expression of FAMA, which controls the final stages of differentiation, promoting guard cell (GC) identity acquisition and the irreversible termination of the meristematic activity of the cells (Figure 2C) [99][100][101]. AtRBR plays important roles in the regulatory network of stomata development, and its downregulation at the GMC or GC stages, induces extra divisions in differentiated GCs and the formation of aberrant stomata-in-stomatal nested structures [102][101]. In fact, AtRBR hyperphosphorylation inhibits stomatal initiation affecting the asymmetric division of protodermal cells that produces MMCs, this seems to be controlled by CDKA;1, that negatively regulates AtRBR, and regulates positively SPCH TF through phosphorylation (Figure 2C) [103][104][105]. It has also been hypothesized that AtRBR hyperphosphorylation by CDKB1;1-CYCD7;1 inhibits the AtRBR/FAMA repression complex leading to the induction of cell-cycle regulators of the GMC symmetric division event [106][107][108]. At the same time, MUTE directly upregulates FAMA and FLP; and FAMA represses cell-cycle control genes such as CDKB1;1, ensuring a single symmetric division to form GCs (Figure 2C) [109][99]. Mutation in the FAMA LxCxE sequence prevents the formation of the AtRBR/FAMA complex, making cells unable to maintain the long-term commitment to differentiate into GC, and arresting this process at the GMC stage [101][106]. Finally, it is also possible that FAMA functions at early steps of guard cell differentiation since the AtRBR/FAMA heterodimer binds to SPCH and FAMA promoters, and this complex negatively regulates the accumulation of the SPCH transcript, which is normally expressed at early stages of guard cells development (Figure 2C) [101][110].
As it can be appreciated, the differentiation processes of skeletal muscle in mammals and guard cells in Arabidopsis are both regulated by a similar set of conserved elements: pRb/RBR, bHLHs, cyclins and CDKs (Figure 2D).
Function of pRb in Epigenetic Modifications, Chromatin Regulation, and DNA Damage
In mammals and plants, many epigenetic modifier proteins interact with protein complexes that include pRb/RBR, allowing them to regulate different developmental processes.
Human pRb has been reported to interact with over 300 proteins and many such protein interactions are epigenetic modifier proteins or interact with the latter [111][112]. pRb levels decrease leads to an incomplete chromosome condensation and segregation during mitosis, as it has been observed in cancer cells; some alterations of the chromatin structure are also induced by changes in histone methylation and acetylation [113][114][115]. pRb can interact with chromatin remodeling factors, such as histone deacetylases, DNA methyltransferases, histone methyltransferases, and with complexes like SWI/SNF; and like Polycomb group (PcG), the latter is a chromatin-modifying complex that maintains repressed gene expression states and is subdivided into two main complexes: Polycomb repressive complex 1 (PRC1) and PRC2 [113][116][117][118][119].
In the case of a double strand break, pRb is necessary to form the complex of the heterodimer E2F1-pRb with TopBP1 (DNA topoisomerase 2-binding protein 1) [120]. TopBP1 is a protein that interacts with Topoisomerase 2β (Top2β) and with other proteins that participate in DNA replication and in the maintenance of the DNA integrity and genome stability [115][121]. In addition, the protein complex E2F1-pRb-TopBP1 interacts with BRG1 (also known as ATP-dependent chromatin remodeler SMARCA4) which is a member of the SWI/SNF complex that is necessary to reduce nucleosome density at injury sites, allowing DNA end resection and reparation by homologous recombination (HR) [120][122]. pRb interacts with the tumor suppressor BRCA1 (Breast cancer 1), which is also involved in DNA repair via Homologous Recombination. The BRCA1-pRb complex interacts with histone deacetylases (HDAC1/2) and with topoisomerase 2β (Top2β) to regulate DNA stability [123][124].
In plants, there are also numerous reports of RBR interactions with epigenetic modifiers, which are important in the regulation of different developmental processes [125]. In Arabidopsis, like in animals, it has been shown that PRC2, a subcomplex of PcG, participates together with AtRBR in the establishment of the H3K27me3 mark during differentiation and development of the female and male gametophytes, in leaf development and during the establishment of stoma cell lineages. In these three processes, AtRBR associates with components of the PRC2 repressor complex such as MULTICOPYSUPPRESSOR OF IRA1 (MSI1), FERTILIZATION INDEPENDENT ENDOSPERM (FIE), VERNALIZATION 2 (VRN2), and CURLY LEAF (CLF), a gene orthologous to EZH2 from humans [126][127]. In addition, AtRBR together with MSI1 directly represses the expression of the DNA methylase METHYLTRANSFERASE 1 (MET1), that maintains DNA methylation during DNA replication and regulates gene imprinting. The repression of MET1 by this complex allows the transcriptional activation of FERTILIZATION INDEPENDENT SEED 2 (FIS2) and FLOWERING WAGENINGEN (FWA), that are important for female gametogenesis [127][128][129]. In plant embryos, the PRC2 complex with AtRBR directly binds and deposits the H2K27m3 mark on different embryonic genes, leading to their repression and subsequent seed germination [130][131].
AtRBR also appears to regulate DNA repair in several conditions. TOP1α is critical to ensure genome integrity and survival of root stele stem cells, as the loss of function of TOP1α triggers DNA double-strand breaks and cell death in these cells; in the root, TOP1α is downregulated by AtRBR [132]. Although the participation of AtRBR and Top1α in the shoot meristem have not yet been studied, TOP1α participates with the PRC2 complex in the repression of the WUS locus [133][132][134].
AtRBR also is recruited to damaged DNA sites, along with E2Fa and AtBRCA1 and helps to maintain the integrity of the root meristem. Furthermore, similar to what is observed for animals for BRCA1 and pRb [116][135], AtRBR and AtBRCA1 have been shown to physically interact when cells are damaged [136]. In addition, E2Fa is required for AtBRCA1 expression, when genotoxic stress is induced [137]. Finally, analysis of chromosome sites to which AtRBR physically binds, show that this protein not only targets gene regulatory sequences, but also transposons, especially Miniature Inverted-repeat Transposable Elements (MITEs) [57].
This entry is adapted from the peer-reviewed paper 10.3390/ijms21144925
References
- Baldi, A.; Boccia, V.; Claudio, P.P.; De Luca, A.; Giordano, A.; Genomic structure of the human retinoblastoma-related Rb2/p130 gene.. Proceedings of the National Academy of Sciences of the United States of America 1996, 93, 4629–4632, , doi:10.1073/pnas.93.10.4629..
- Claudio, P.P.; Tonini, T.; Giordano, A.; The retinoblastoma family: Twins or distant cousins?. Genome Biology 2002, 3, 3012–3012, doi:10.1186/gb-2002-3-9-reviews3012..
- Ichimura, K.; Hanafusa, H.; Takimoto, H.; Ohgama, Y.; Akagi, T.; Shimizu, K.; Structure of the human retinoblastoma-related p107 gene and its intragenic deletion in a B-cell lymphoma cell line.. Gene 2000, 251 , 37–43 , doi:10.1016/S0378-1119(00)00193-1.
- Chinnam, M.; Goodrich, D.W.; RB1, Development, and Cancer.. Current Topics in Developmental Biology 2011, 94, 129–169, doi:10.1016/B978-0-12-380916-2.00005-X.
- Dick, F.A.; Structure-function analysis of the retinoblastoma tumor suppressor protein – is the whole a sum of its parts?. Cell Division 2007, 2, 26 , doi:10.1186/1747-1028-2-26.
- Dick, F.A.; Rubin, S.M.; Molecular mechanisms underlying RB protein function.. Nature Reviews Molecular Cell Biology 2013, 14, 297–306, doi:10.1038/nrm3567.
- Lee, J.O.; Russo, A.A.; Pavletich, N.P.; Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7.. Nature 1998, 391, 859–865, doi:10.1038/36038.
- Morris, E.J.; Dyson, N.J.; Retinoblastoma protein partners.. Advances in Cancer Research 2001, 82, 1–54, , doi:10.1016/S0065-230X(01)82001-7.
- Sanidas, I.; Morris, R.; Fella, K.A.; Rumde, P.H.; Boukhali, M.; Tai, E.C.; Ting, D.T.; Lawrence, M.S.; Haas, W.; Dyson, N.J.; et al. A Code of Mono-phosphorylation Modulates the Function of RB.. Molecular Cell 2019, 73, 985-1000.e6, , doi:10.1016/j.molcel.2019.01.004.
- Sanidas, I.; Morris, R.; Fella, K.A.; Rumde, P.H.; Boukhali, M.; Tai, E.C.; Ting, D.T.; Lawrence, M.S.; Haas, W.; Dyson, N.J.; et al. A Code of Mono-phosphorylation Modulates the Function of RB.. Molecular Cell 2019, 73, 985-1000.e6, , doi:10.1016/j.molcel.2019.01.004.
- Chan, H.M.; Krstic-Demonacos, M.; Smith, L.; Demonacos, C.; La Thangue, N.B.; Acetylation control of the retinoblastoma tumour-suppressor protein.. Nature Cell Biology 2001, 3, 667–674, doi:10.1038/35083062.
- Pickard, A.; Wong, P.P.; McCance, D.J.; Acetylation of Rb by PCAF is required for nuclear localization and keratinocyte differentiation. . Journal of Cell Science 2010, 123, 3718–3726, doi:10.1242/jcs.068924.
- Kim, K.Y.; Wang, D.-H.; Campbell, M.; Huerta, S.B.; Shevchenko, B.; Izumiya, C.; Izumiya, Y.; PRMT4-mediated arginine methylation negatively regulates retinoblastoma tumor suppressor protein and promotes E2F-1 dissociation.. Molecular and cellular biology 2015, 35, 238–48, doi:10.1128/MCB.00945-14.
- Saddic, L.A.; West, L.E.; Aslanian, A.; Yates, J.R.; Rubin, S.M.; Gozani, O.; Sage, J.; Methylation of the retinoblastoma tumor suppressor by SMYD2.. Journal of Biological Chemistry 2010, 285, 37733–37740, doi:10.1074/jbc.M110.137612.
- Munro, S.; Khaire, N.; Inche, A.; Carr, S.; La Thangue, N.B.; Lysine methylation regulates the pRb tumour suppressor protein.. Oncogene 2010, 29, 2357–2367, doi:10.1038/onc.2009.511.
- Ledl, A.; Schmidt, D.; Müller, S.; Viral oncoproteins E1A and E7 and cellular LxCxE proteins repress SUMO modification of the retinoblastoma tumor suppressor. . Oncogene 2005, 24, 3810–3818, doi:10.1038/sj.onc.1208539.
- Meng, F.; Qian, J.; Yue, H.; Li, X.; Xue, K.; SUMOylation of Rb enhances its binding with CDK2 and phosphorylation at early G1 phase. . Cell Cycle 2016, 15, 1724–1732, doi:10.1080/15384101.2016.1182267.
- Miwa, S.; Uchida, C.; Kitagawa, K.; Hattori, T.; Oda, T.; Sugimura, H.; Yasuda, H.; Nakamura, H.; Chida, K.; Kitagawa, M.; et al. Mdm2-mediated pRB downregulation is involved in carcinogenesis in a p53-independent manner. . Biochemical and Biophysical Research Communications 2006, 340, 54–61, doi:10.1016/j.bbrc.2005.11.148.
- Friend, S.H.; Bernards, R.; Rogelj, S.; Weinberg, R.A.; Rapaport, J.M.; Albert, D.M.; Dryja, T.P.; A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. . Nature 1986, 323, 643–646, doi:10.1038/323643a0.
- McGee, T.L.; Yandell, D.W.; Dryja, T.P.; Structure and partial genomic sequence of the human retinoblastoma susceptibility gene. . Gene 1989, 80, 119–128, doi:10.1016/0378-1119(89)90256-4.
- Hong, F.D.; Huang, H.J.S.; To, H.; Young, L.J.S.; Oro, A.; Bookstein, R.; Lee, E.Y.H.P.; Lee, W.H.; Structure of the human retinoblastoma gene. . Proceedings of the National Academy of Sciences of the United States of America 1989, 86, 5502–5506, doi:10.1073/pnas.86.14.5502.
- Shen, B.; Carneiro, N.; Torres-Jerez, I.; Stevenson, B.; McCreery, T.; Helentjaris, T.; Baysdorfer, C.; Almira, E.; Ferl, R.J.; Habben, J.E.; et al. Partial sequencing and mapping of clones from two maize cDNA libraries. . Plant molecular biology 1994, 26, 1085–1101, doi:10.1007/bf00040691.
- Xie, Q.; Sanz-Burgos, A.P.; Hannon, G.J.; Gutiérrez, C.; Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. . The EMBO Journal 1996, 15, 4900–4908, doi:10.1002/j.1460-2075.1996.tb00870.x.
- Xie, Q.; Suárez-López, P.; Gutiérrez, C.; Identification and analysis of a retinoblastoma binding motif in the replication protein of a plant DNA virus: requirement for efficient viral DNA replication. . The EMBO Journal 1995, 14, 4073–4082, doi:10.1002/j.1460-2075.1995.tb00079.x.
- Nakagami, H.; Sekine, M.; Murakami, H.; Shinmyo, A.; Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. . The Plant journal : for cell and molecular biology 1999, 18, 243–252, doi:10.1046/j.1365-313x.1999.00449.x.
- Kong, L.-J.; A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. . The EMBO Journal 2000, 19, 3485–3495, doi:10.1093/emboj/19.13.3485.
- Lendvai, A.; Pettko-Szandtner, A.; Csordas-Toth, E.; Miskolczi, P.; Horvath, G. V.; Gyorgyey, J.; Dudits, D.; Dicot and monocot plants differ in retinoblastoma-related protein subfamilies. . Journal of Experimental Botany 2007, 58, 1663–1675, doi:10.1093/jxb/erm022.
- Durfee, T.; Feiler, H.S.; Gruissem, W.; Retinoblastoma-related proteins in plants: homologues or orthologues of their metazoan counterparts? . Plant Molecular Biology 2000, 43, 635–642, doi:10.1023/A:1006426808185.
- Kuwabara, A.; Gruissem, W.; Arabidopsis Retinoblastoma-related and Polycomb group proteins: Cooperation during plant cell differentiation and development. . Journal of Experimental Botany 2014, 65, 2667–2676, doi:10.1093/jxb/eru069.
- De Jager, S.M.; Murray, J.A.H.; Retinoblastoma proteins in plants. . Plant Molecular Biology 1999, 41, 295–299, doi:10.1023/A:1006398232003.
- Desvoyes, B.; De Mendoza, A.; Ruiz-Trillo, I.; Gutierrez, C.; Novel roles of plant RETINOBLASTOMA-RELATED (RBR) protein in cell proliferation and asymmetric cell division. . Journal of Experimental Botany 2014, 65, 2657–2666, doi:10.1093/jxb/ert411.
- Gutzat, R.; Borghi, L.; Gruissem, W.; Emerging roles of RETINOBLASTOMA-RELATED proteins in evolution and plant development. . Trends in Plant Science 2012, 17, 139–148, doi:10.1016/j.tplants.2011.12.001.
- Reiland, S.; Messerli, G.; Baerenfaller, K.; Gerrits, B.; Endler, A.; Grossmann, J.; Gruissem, W.; Baginsky, S.; Large-scale arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks1[w]. . Plant Physiology 2009, 150, 889–903 , doi:10.1104/pp.109.138677.
- Grant, G.D.; Cook, J.G.; The temporal regulation of s phase proteins during G1. . Advances in Experimental Medicine and Biology 2017, 1042, 335–369, doi:10.1007/978-981-10-6955-0_16.
- Takaki, T.; Fukasawa, K.; Suzuki-Takahashi, I.; Hirai, H.; Cdk-mediated phosphorylation of pRB regulates HDAC binding in vitro. . Biochemical and Biophysical Research Communications 2004, 316, 252–255, doi:10.1016/j.bbrc.2004.02.044.
- Scott, M.C.; Sarver, A.L.; Tomiyasu, H.; Cornax, I.; Van Etten, J.; Varshney, J.; O’Sullivan, M.G.; Subramanian, S.; Modiano, J.F.; Aberrant retinoblastoma (RB)-E2F transcriptional regulation defines molecular phenotypes of osteosarcoma. . Journal of Biological Chemistry 2015, 290, 28070–28083, doi:10.1074/jbc.M115.679696.
- Dowdy, S.F.; Hinds, P.W.; Louie, K.; Reed, S.I.; Arnold, A.; Weinberg, R.A.; Physical interaction of the retinoblastoma protein with human D cyclins. . Cell 1993, 73, 499–511, doi:10.1016/0092-8674(93)90137-f.
- Kato, J.; Matsushime, H.; Hiebert, S.W.; Ewen, M.E.; Sherr, C.J.; Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. . Genes and Development 1993, 7, 331–342, doi:10.1101/gad.7.3.331.
- Gillett, C.; Fantl, V.; Smith, R.; Fisher, C.; Bartek, J.; Dickson, C.; Barnes, D.; Peters, G.; Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. . Cancer research 1994, 54, 1812–7., .
- Rizzolio, F.; Tuccinardi, T.; Caligiuri, I.; Lucchetti, C.; Giordano, A.; CDK Inhibitors: From the Bench to Clinical Trials. . Current Drug Targets 2010, 11, 279–290, doi:10.2174/138945010790711978.
- Markey, M.P.; Bergseid, J.; Bosco, E.E.; Stengel, K.; Xu, H.; Mayhew, C.N.; Schwemberger, S.J.; Braden, W.A.; Jiang, Y.; Babcock, G.F.; et al. Loss of the retinoblastoma tumor suppressor: Differential action on transcriptional programs related to cell cycle control and immune function. . Oncogene 2007, 26, 6307–6318, doi:10.1038/sj.onc.1210450.
- Fischer, M.; Müller, G.A.; Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes. . Critical Reviews in Biochemistry and Molecular Biology 2017, 52, 638–662, doi:10.1080/10409238.2017.1360836..
- Sadasivam, S.; DeCaprio, J.A.; The DREAM complex: master coordinator of cell cycle-dependent gene expression. . Nature Reviews Cancer 2013, 13, 585–595, doi:10.1038/nrc3556.
- Guiley, K.Z.; Liban, T.J.; Felthousen, J.G.; Ramanan, P.; Litovchick, L.; Rubin, S.M.; Structural mechanisms of DREAM complex assembly and regulation. . Genes and Development 2015, 29, 961–974, doi:10.1101/gad.257568.114..
- Subhashini Sadasivam; Shenghua Duan; James A DeCaprio; The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes & Development 2012, 26, 474-489, 10.1101/gad.181933.111.
- Xie, Q.; Suárez-López, P.; Gutiérrez, C.; Identification and analysis of a retinoblastoma binding motif in the replication protein of a plant DNA virus: requirement for efficient viral DNA replication. . The EMBO Journal 1995, 14, 4073–4082, doi:10.1002/j.1460-2075.1995.tb00079.x..
- Marlis Dahl; Irute Meskiene; László Bögre; Dang Thi Cam Ha; Ines Swoboda; Rainer Hubmann; Heribert Hirt; Erwin Heberle-Bors; The D-Type Alfalfa Cyclin Gene cycMs4 Complements G 1 Cyclin-Deficient Yeast and Is Induced in the G 1 Phase of the Cell Cycle. The Plant Cell 1995, 7, 1847, 10.2307/3870192.
- Rachael P. Huntley; Sandra Healy; Donna Freeman; Paul Lavender; Sarah De Jager; Judith Greenwood; Joe Makker; Edward Walker; Mark Jackman; Qi Xie; et al. The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins.. Plant Molecular Biology 1998, 37, 155-169, 10.1023/a:1005902226256.
- Rajeev Soni; Jeremy P. Carmichael; Zahid Shah; James A. H. Murray; A Family of Cyclin D Homologs from Plants Differentially Controlled by Growth Regulators and Containing the Conserved Retinoblastoma Protein Interaction Motif. The Plant Cell 1994, 7, 85, 10.2307/3869840.
- R A Ach; T Durfee; A B Miller; P Taranto; L Hanley-Bowdoin; P C Zambryski; W Gruissem; RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein.. Molecular and Cellular Biology 1997, 17, 5077-5086, 10.1128/mcb.17.9.5077.
- Maria Boniotti; Crisanto Gutiérrez; A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex.. The Plant Journal 2001, 28, 341-350, 10.1046/j.1365-313X.2001.01160.x.
- Hirofumi Nakagami; Kazue Kawamura; Keiko Sugisaka; Masami Sekine; Atsuhiko Shinmyo; Phosphorylation of Retinoblastoma-Related Protein by the Cyclin D/Cyclin-Dependent Kinase Complex Is Activated at the G1/S-Phase Transition in TobaccoW⃞. The Plant Cell 2002, 14, 1847-1857, 10.1105/tpc.002550.
- Martin Fischer; James A. DeCaprio; Does Arabidopsis thaliana DREAM of cell cycle control?. The EMBO Journal 2015, 34, 1987-1989, 10.15252/embj.201592196.
- Zoltán Magyar; László Bögre; Masaki Ito; DREAMs make plant cells to cycle or to become quiescent. Current Opinion in Plant Biology 2016, 34, 100-106, 10.1016/j.pbi.2016.10.002.
- Kosuke Kobayashi; Toshiya Suzuki; Eriko Iwata; Zoltán Magyar; László Bögre; Masaki Ito; MYB3Rs, plant homologs of Myb oncoproteins, control cell cycle-regulated transcription and form DREAM-like complexes. Transcription 2015, 6, 106-111, 10.1080/21541264.2015.1109746.
- Wanpeng Wang; Paja Sijacic; Pengbo Xu; Hongli Lian; Zhongchi Liu; Arabidopsis TSO1 and MYB3R1 form a regulatory module to coordinate cell proliferation with differentiation in shoot and root. Proceedings of the National Academy of Sciences 2018, 115, E3045-E3054, 10.1073/pnas.1715903115.
- Daniel Bouyer; Maren Heese; PoYu Chen; Hirofumi Harashima; François Roudier; Christian Grüttner; Arp Schnittger; Genome-wide identification of RETINOBLASTOMA RELATED 1 binding sites in Arabidopsis reveals novel DNA damage regulators. PLOS Genetics 2018, 14, e1007797, 10.1371/journal.pgen.1007797.
- Cizhong Jiang; Jianying Gu; Surinder Chopra; Xun Gu; Thomas Peterson; Ordered origin of the typical two- and three-repeat Myb genes. Gene 2004, 326, 13-22, 10.1016/j.gene.2003.09.049.
- Kosuke Kobayashi; Toshiya Suzuki; Eriko Iwata; Norihito Nakamichi; Takamasa Suzuki; PoYu Chen; Misato Ohtani; Takashi Ishida; Hanako Hosoya; Sabine Müller; et al. Transcriptional repression by MYB 3R proteins regulates plant organ growth. The EMBO Journal 2015, 34, 1992-2007, 10.15252/embj.201490899.
- Stuart A. Newman; Cell differentiation: What have we learned in 50 years?. Journal of Theoretical Biology 2019, 485, 110031, 10.1016/j.jtbi.2019.110031.
- Alejandro Sánchez Alvarado; Shinya Yamanaka; Rethinking differentiation: stem cells, regeneration, and plasticity.. Cell 2014, 157, 110-9, 10.1016/j.cell.2014.02.041.
- G De Falco; F Comes; Cristiano Simone; pRb: master of differentiation. Coupling irreversible cell cycle withdrawal with induction of muscle-specific transcription. Oncogene 2006, 25, 5244-5249, 10.1038/sj.onc.1209623.
- David W. Goodrich; The retinoblastoma tumor-suppressor gene, the exception that proves the rule. Oncogene 2006, 25, 5233-5243, 10.1038/sj.onc.1209616.
- Konstantinos E. Hatzistergos; Adam R. Williams; Derek M Dykxhoorn; Michael A. Bellio; Wendou Yu; Joshua M. Hare; Tumor Suppressors RB1 and CDKN2a Cooperatively Regulate Cell-Cycle Progression and Differentiation During Cardiomyocyte Development and Repair. Circulation Research 2019, 124, 1184-1197, 10.1161/circresaha.118.314063.
- L Khidr; Phang-Lang Chen; RB, the conductor that orchestrates life, death and differentiation. Oncogene 2006, 25, 5210-5219, 10.1038/sj.onc.1209612.
- Jingling Li; Cyndhavi Narayanan; Jing Bian; Danielle Sambo; Thomas Brickler; Wancong Zhang; Sundari Chetty; A transient DMSO treatment increases the differentiation potential of human pluripotent stem cells through the Rb family. PLOS ONE 2018, 13, e0208110, 10.1371/journal.pone.0208110.
- Marta M. Lipinski; Tyler Jacks; The retinoblastoma gene family in differentiation and development. Oncogene 1999, 18, 7873-7882, 10.1038/sj.onc.1203244.
- Marjolein Wildwater; Ana Campilho; José Manuel Pérez-Pérez; Renze Heidstra; Ikram Blilou; Henrie Korthout; Jayanta Chatterjee; Luisa Mariconti; Wilhelm Gruissem; Ben Scheres; et al. The RETINOBLASTOMA-RELATED Gene Regulates Stem Cell Maintenance in Arabidopsis Roots. Cell 2005, 123, 1337-1349, 10.1016/j.cell.2005.09.042.
- Bénédicte Desvoyes; Elena Ramirez-Parra; Qi Xie; Nam-Hai Chua; Crisanto Gutiérrez; Cell Type-Specific Role of the Retinoblastoma/E2F Pathway during Arabidopsis Leaf Development1. Plant Physiology 2005, 140, 67-80, 10.1104/pp.105.071027.
- Joanna Wyrzykowska; Martine Schorderet; Stéphane Pien; Wilhelm Gruissem; Andrew Fleming; Induction of Differentiation in the Shoot Apical Meristem by Transient Overexpression of a Retinoblastoma-Related Protein. PLANT PHYSIOLOGY 2006, 141, 1338-1348, 10.1104/pp.106.083022.
- Xin’Ai Zhao; Jonathan Bramsiepe; Matthias Van Durme; Shinichiro Komaki; Maria Ada Prusicki; Daisuke Maruyama; Joachim Forner; Anna Medzihradszky; Erik Wijnker; Hirofumi Harashima; et al. RETINOBLASTOMA RELATED1 mediates germline entry in Arabidopsis. Science 2017, 356, eaaf6532, 10.1126/science.aaf6532.
- Serena Perilli; Jose Manuel Perez-Perez; Riccardo Di Mambro; Cristina Llavata Peris; Sara Diaz-Trivino; Marta Del Bianco; Emanuela Pierdonati; Laila Moubayidin; Alfredo Cruz-Ramírez; Paolo Costantino; et al. RETINOBLASTOMA-RELATED Protein Stimulates Cell Differentiation in the Arabidopsis Root Meristem by Interacting with Cytokinin Signaling[W]. The Plant Cell 2013, 25, 4469-4478, 10.1105/tpc.113.116632.
- Aarthi Putarjunan; Keiko U. Torii; Stomagenesis versus myogenesis: Parallels in intrinsic and extrinsic regulation of transcription factor mediated specialized cell-type differentiation in plants and animals. Development, Growth & Differentiation 2016, 58, 341-354, 10.1111/dgd.12282.
- Dominique C Bergmann; Juliana L. Matos; Convergence of stem cell behaviors and genetic regulation between animals and plants: insights from the Arabidopsis thaliana stomatal lineage. F1000Prime Reports 2014, 6, 53, 10.12703/p6-53.
- Fabien Le Grand; Michael A Rudnicki; Skeletal muscle satellite cells and adult myogenesis. Current Opinion in Cell Biology 2007, 19, 628-633, 10.1016/j.ceb.2007.09.012.
- Bennett G. Novitch; G J Mulligan; T Jacks; A B Lassar; Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle.. Journal of Cell Biology 1996, 135, 441-456, 10.1083/jcb.135.2.441.
- Peter S. Zammit; Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis.. Seminars in Cell & Developmental Biology 2017, 72, 19-32, 10.1016/j.semcdb.2017.11.011.
- Ddw Cornelison; Bradley B. Olwin; Michael A. Rudnicki; Barbara J. Wold; MyoD−/− Satellite Cells in Single-Fiber Culture Are Differentiation Defective and MRF4 Deficient. Developmental Biology 2000, 224, 122-137, 10.1006/dbio.2000.9682.
- Takeshi Endo; Susumu Goto; Retinoblastoma Gene Product Rb Accumulates during Myogenic Differentiation and Is Deinduced by the Expression of SV40 Large T Antigen1. Journal of Biochemistry 1992, 112, 427-430, 10.1093/oxfordjournals.jbchem.a123916.
- Michael S. Huh; Maura H. Parker; Anthony Scimè; Robin Parks; Michael A. Rudnicki; Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. Journal of Cell Biology 2004, 166, 865-876, 10.1083/jcb.200403004.
- Stephen J. Tapscott; The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 2005, 132, 2685-2695, 10.1242/dev.01874.
- Wei Gu; Jay W. Schneider; Gianluigi Condorelli; Sunjay Kaushal; Vijak Mahdavi; Bernardo Nadal-Ginard; Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell 1993, 72, 309-324, 10.1016/0092-8674(93)90110-c.
- Pawel Smialowski; Mahavir Singh; Aleksandra Mikolajka; Sudipta Majumdar; Joma Kanikadu Joy; Narasimharao Nalabothula; Marcin Krajewski; Roland Degenkolbe; Hans-Ulrich Bernard; Tad A. Holak; et al. NMR and mass spectrometry studies of putative interactions of cell cycle proteins pRb and CDK6 with cell differentiation proteins MyoD and ID-2. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 2005, 1750, 48-60, 10.1016/j.bbapap.2005.03.012.
- Alessandra Magenta; Carlo Cenciarelli; Francesca De Santa; Paola Fuschi; Fabio Martelli; Maurizia Caruso; Armando Felsani; MyoD Stimulates RB Promoter Activity via the CREB/p300 Nuclear Transduction Pathway. Molecular and Cellular Biology 2003, 23, 2893-2906, 10.1128/mcb.23.8.2893-2906.2003.
- Martelli, F.; Cenciarelli, C.; Santarelli, G.; Polikar, B.; Felsani, A.; Caruso, M.; MyoD induces retinoblastoma gene expression during myogenic differentiation. . Oncogene 1994, 9, 3579–3590, .
- James Dr Knight; Rashmi Kothary; The myogenic kinome: protein kinases critical to mammalian skeletal myogenesis. Skeletal Muscle 2011, 1, 29-29, 10.1186/2044-5040-1-29.
- Hasan N. Rajabi; Chiaki Takahashi; Mark E. Ewen; Anja M. Swenson; Darshan V. Trivedi; Anna A. Rauscher; Yuan Wang; Yasuharu Takagi; Bradley M. Palmer; András Málnási-Csizmadia; et al. Retinoblastoma Protein and MyoD Function Together to Effect the Repression of Fra-1 and in Turn Cyclin D1 during Terminal Cell Cycle Arrest Associated with Myogenesis. Journal of Biological Chemistry 2014, 289, 23417-23427, 10.1074/jbc.m113.532572.
- S X Skapek; J Rhee; P S Kim; Bennett G. Novitch; A B Lassar; Cyclin-mediated inhibition of muscle gene expression via a mechanism that is independent of pRB hyperphosphorylation.. Molecular and Cellular Biology 1996, 16, 7043-7053, 10.1128/mcb.16.12.7043.
- Suzan Ruijtenberg; Sander Van Den Heuvel; Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression.. Cell Cycle 2015, 15, 196-212, 10.1080/15384101.2015.1120925.
- An Song; Qi Wang; Mark G. Goebl; Maureen A. Harrington; Phosphorylation of Nuclear MyoD Is Required for Its Rapid Degradation. Molecular and Cellular Biology 1998, 18, 4994-4999, 10.1128/mcb.18.9.4994.
- Jian-Min Zhang; Qin Wei; Xiaohang Zhao; Bruce M. Paterson; Coupling of the cell cycle and myogenesis through the cyclin D1-dependent interaction of MyoD with cdk4.. The EMBO Journal 1999, 18, 926-933, 10.1093/emboj/18.4.926.
- Pier Lorenzo Puri; Simona Iezzi; Peter Stiegler; Tung-Ti Chen; R.Louis Schiltz; George Muscat; Antonio Giordano; Larry Kedes; J Y Wang; Vittorio Sartorelli; et al. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis.. Molecular Cell 2001, 8, 885-897, 10.1016/s1097-2765(01)00373-2.
- Cristiano Simone; Sonia-V. Forcales; David A Hill; Anthony N Imbalzano; Lucia Latella; Pier Lorenzo Puri; p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nature Genetics 2004, 36, 738-743, 10.1038/ng1378.
- Lynn J. Pillitteri; Juan Dong; Stomatal Development in Arabidopsis. The Arabidopsis Book 2013, 11, e0162, 10.1199/tab.0162.
- Anne Vatén; Dominique C Bergmann; Mechanisms of stomatal development: an evolutionary view. EvoDevo 2012, 3, 11-11, 10.1186/2041-9139-3-11.
- Julie E. Gray; Plant Development: Three Steps for Stomata. Current Biology 2007, 17, R213-R215, 10.1016/j.cub.2007.01.032.
- Masahiro M. Kanaoka; Lynn Jo Pillitteri; Hiroaki Fujii; Yuki Yoshida; Naomi L. Bogenschutz; Junji Takabayashi; Jian-Kang Zhu; Keiko U. Torii; SCREAM/ICE1 and SCREAM2 Specify Three Cell-State Transitional Steps Leading to Arabidopsis Stomatal Differentiation. The Plant Cell 2008, 20, 1775-1785, 10.1105/tpc.108.060848.
- Lynn Jo Pillitteri; Keiko U. Torii; Breaking the silence: three bHLH proteins direct cell-fate decisions during stomatal development. BioEssays 2007, 29, 861-870, 10.1002/bies.20625.
- Soon-Ki Han; Xingyun Qi; Kei Sugihara; Jonathan H. Dang; Takaho A. Endo; Kristen L. Miller; Eun-Deok Kim; Takashi Miura; Keiko U. Torii; MUTE Directly Orchestrates Cell-State Switch and the Single Symmetric Division to Create Stomata. Developmental Cell 2018, 45, 303-315.e5, 10.1016/j.devcel.2018.04.010.
- Nicholas Zoulias; Emily L. Harrison; Stuart A. Casson; Julie E. Gray; Molecular control of stomatal development. Biochemical Journal 2018, 475, 441-454, 10.1042/bcj20170413.
- Juliana L Matos; On Sun Lau; Charles Hachez; Alfredo Cruz-Ramírez; B. Scheres; Dominique C Bergmann; Irreversible fate commitment in the Arabidopsis stomatal lineage requires a FAMA and RETINOBLASTOMA-RELATED module. eLife 2014, 3, 1–15, 10.7554/eLife.03271.
- Lorenzo Borghi; Ruben Gutzat; Johannes Fütterer; Yec'han Laizet; Lars Hennig; Wilhelm Gruissem; Arabidopsis RETINOBLASTOMA-RELATED Is Required for Stem Cell Maintenance, Cell Differentiation, and Lateral Organ Production. The Plant Cell 2010, 22, 1792-1811, 10.1105/tpc.110.074591.
- Soon-Ki Han; Keiko U. Torii; Linking cell cycle to stomatal differentiation. Current Opinion in Plant Biology 2019, 51, 66-73, 10.1016/j.pbi.2019.03.010.
- Ke-Zhen Yang; Min Jiang; Ming Wang; Shan Xue; Ling-Ling Zhu; Hong-Zhe Wang; Jun-Jie Zou; Eun-Kyoung Lee; Fred Sack; Jie Le; et al. Phosphorylation of Serine 186 of bHLH Transcription Factor SPEECHLESS Promotes Stomatal Development in Arabidopsis. Molecular Plant 2015, 8, 783-795, 10.1016/j.molp.2014.12.014.
- Moritz K. Nowack; Hirofumi Harashima; Nico Dissmeyer; Xin'ai Zhao; Daniel Bouyer; Annika Weimer; Freya De Winter; Fang Yang; Arp Schnittger; Genetic Framework of Cyclin-Dependent Kinase Function in Arabidopsis. Developmental Cell 2012, 22, 1030-1040, 10.1016/j.devcel.2012.02.015.
- Eunkyoung Lee; Jessica Regan Lucas; Fred Sack; Deep functional redundancy between FAMA and FOUR LIPS in stomatal development. The Plant Journal 2014, 78, 555-565, 10.1111/tpj.12489.
- Annika K. Weimer; Juliana L. Matos; Nidhi Sharma; Farah Patell; James A.H. Murray; Walter Dewitte; Dominique C. Bergmann; Lineage- and stage-specific expressed CYCD7;1 coordinates the single symmetric division that creates stomatal guard cells. Development 2018, 145, dev160671, 10.1242/dev.160671.
- Jorge Zamora-Zaragoza; B. Scheres; Tuning Division and Differentiation in Stomata: How to Silence a MUTE.. Developmental Cell 2018, 45, 282-283, 10.1016/j.devcel.2018.04.019.
- Charles Hachez; Kyoko Ohashi-Ito; Juan Dong; Dominique C. Bergmann; Differentiation of Arabidopsis Guard Cells: Analysis of the Networks Incorporating the Basic Helix-Loop-Helix Transcription Factor, FAMA. Plant Physiology 2011, 155, 1458-1472, 10.1104/pp.110.167718.
- Liang Chen; Zhongliang Wu; Suiwen Hou; SPEECHLESS Speaks Loudly in Stomatal Development.. Frontiers in Plant Science 2020, 11, 114, 10.3389/fpls.2020.00114.
- Ioannis Sanidas; Robert Morris; Katerina A. Fella; Purva H. Rumde; Myriam Boukhali; Eric C. Tai; David T. Ting; Michael S. Lawrence; Wilhelm Haas; Nicholas J. Dyson; et al. A Code of Mono-phosphorylation Modulates the Function of RB. Molecular Cell 2019, 73, 985-1000.e6, 10.1016/j.molcel.2019.01.004.
- Francesco Paolo Fiorentino; Irene Marchesi; Antonio Giordano; On the role of retinoblastoma family proteins in the establishment and maintenance of the epigenetic landscape. Journal of Cellular Physiology 2012, 228, 276-284, 10.1002/jcp.24141.
- C Uchida; Roles of pRB in the Regulation of Nucleosome and Chromatin Structures. BioMed Research International 2016, 2016, 1-11, 10.1155/2016/5959721.
- Susana Gonzalo; Maria A. Blasco; Role of Rb Family in the Epigenetic Definition of Chromatin. Cell Cycle 2005, 4, 752-755, 10.4161/cc.4.6.1720.
- Renier Velez-Cruz; D. G. Johnson; The Retinoblastoma (RB) Tumor Suppressor: Pushing Back against Genome Instability on Multiple Fronts. International Journal of Molecular Sciences 2017, 18, 1776, 10.3390/ijms18081776.
- Olga N. Aprelikova; Bruno S. Fang; Eric G. Meissner; Shane Cotter; Mel Campbell; Aalok Kuthiala; Mika Bessho; Roy A. Jensen; Edison T. Liu; BRCA1-associated growth arrest is RB-dependent. Proceedings of the National Academy of Sciences 1999, 96, 11866-11871, 10.1073/pnas.96.21.11866.
- L. Magnaghi-Jaulin; Sonia Groisman; I. Naguibneva; P. Robin; Stéphanie Lorain; J. P. Le Villain; F. Troalen; Didier Trouche; Annick Harel-Bellan; Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 1998, 391, 601-605, 10.1038/35410.
- Keith D. Robertson; Slimane Ait-Si-Ali; Tomoki Yokochi; Paul A Wade; Peter L. Jones; Alan P. Wolffe; DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nature Genetics 2000, 25, 338-342, 10.1038/77124.
- Bernd Schuettengruber; Henri-Marc Bourbon; Luciano Di Croce; Giacomo Cavalli; Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34-57, 10.1016/j.cell.2017.08.002.
- Renier Velez-Cruz; Swarnalatha Manickavinayaham; Anup K. Biswas; Regina Weaks Clary; Tolkappiyan Premkumar; Francesca Cole; David G. Johnson; RB localizes to DNA double-strand breaks and promotes DNA end resection and homologous recombination through the recruitment of BRG1. Genes & Development 2016, 30, 2500-2512, 10.1101/gad.288282.116.
- Miiko Sokka; Sinikka Parkkinen; Helmut Pospiech; Juhani E. Syväoja; Function of TopBP1 in Genome Stability. Membrane Biogenesis 2009, 50, 119-141, 10.1007/978-90-481-3471-7_7.
- Yuning Jiang; Wai Kit Chu; Potential Roles of the Retinoblastoma Protein in Regulating Genome Editing. Frontiers in Cell and Developmental Biology 2018, 6, 0, 10.3389/fcell.2018.00081.
- Hai Xiao; David W Goodrich; The retinoblastoma tumor suppressor protein is required for efficient processing and repair of trapped topoisomerase II-DNA-cleavable complexes. Oncogene 2005, 24, 8105-8113, 10.1038/sj.onc.1208958.
- Ronit I. Yarden; Lawrence C. Brody; BRCA1 interacts with components of the histone deacetylase complex. Proceedings of the National Academy of Sciences 1999, 96, 4983-4988, 10.1073/pnas.96.9.4983.
- Bénédicte Desvoyes; Marãa Fernã¡ndez-Marcos; Joana Sequeira-Mendes; Sofãa Otero; Zaida Vergara; Crisanto Gutierrez; Maria Fernandez-Marcos; Looking at plant cell cycle from the chromatin window. Frontiers in Plant Science 2014, 5, 369, 10.3389/fpls.2014.00369.
- A. Kuwabara; W Gruissem; Arabidopsis Retinoblastoma-related and Polycomb group proteins: cooperation during plant cell differentiation and development. Journal of Experimental Botany 2014, 65, 2667-2676, 10.1093/jxb/eru069.
- María De La Paz Sanchez; Pamela Aceves-García; Emilio Petrone; Stefan Steckenborn; Rosario Vega-León; Elena R. Alvarez‐Buylla; Adriana Garay; Berenice García-Ponce; The impact of Polycomb group (PcG) and Trithorax group (TrxG) epigenetic factors in plant plasticity. New Phytologist 2015, 208, 684-694, 10.1111/nph.13486.
- Amal J Johnston; Elena Matveeva; Olga Kirioukhova; Ueli Grossniklaus; W Gruissem; A Dynamic Reciprocal RBR-PRC2 Regulatory Circuit Controls Arabidopsis Gametophyte Development. Current Biology 2008, 18, 1680-1686, 10.1016/j.cub.2008.09.026.
- Pauline E Jullien; Assaf Mosquna; Mathieu Ingouff; Tadashi Sakata; Nir Ohad; Frédéric Berger; Retinoblastoma and Its Binding Partner MSI1 Control Imprinting in Arabidopsis. PLOS Biology 2008, 6, e194, 10.1371/journal.pbio.0060194.
- Ruben Gutzat; Lorenzo Borghi; Johannes Fütterer; Sylvain Bischof; Yec‘Han Laizet; Lars Hennig; Regina Feil; John Lunn; Wilhelm Gruissem; RETINOBLASTOMA-RELATED PROTEIN controls the transition to autotrophic plant development. Development 2011, 138, 2977-2986, 10.1242/dev.060830.
- Ruben Gutzat; Lorenzo Borghi; W Gruissem; Emerging roles of RETINOBLASTOMA-RELATED proteins in evolution and plant development. Trends in Plant Science 2012, 17, 139-148, 10.1016/j.tplants.2011.12.001.
- Yonghong Zhang; Lanlan Zheng; Jing Han Hong; Ximing Gong; Chun Zhou; Jose Manuel Perez-Perez; Jian Xu; TOPOISOMERASE1α Acts through Two Distinct Mechanisms to Regulate Stele and Columella Stem Cell Maintenance.. Plant Physiology 2016, 171, 483-93, 10.1104/pp.15.01754.
- Fang Zhang; Kristi Rothermund; Sajithlal B. Gangadharan; Yves Pommier; Edward V. Prochownik; John S. Lazo; Phenotypic Screening Reveals Topoisomerase I as a Breast Cancer Stem Cell Therapeutic Target. Oncotarget 2012, 3, 998-1010, 10.18632/oncotarget.632.
- Xigang Liu; Lei Gao; Thanh Theresa Dinh; Ting Shi; Dongming Li; Ruozhong Wang; Lin Guo; Langtao Xiao; Xuemei Chen; DNA Topoisomerase I Affects Polycomb Group Protein-Mediated Epigenetic Regulation and Plant Development by Altering Nucleosome Distribution in Arabidopsis. The Plant Cell 2014, 26, 2803-2817, 10.1105/tpc.114.124941.
- Serena L. Clark; Ana M. Rodriguez; Russell R. Snyder; Gary D.V. Hankins; Darren Boehning; STRUCTURE-FUNCTION OF THE TUMOR SUPPRESSOR BRCA1. Computational and Structural Biotechnology Journal 2012, 1, e201204005-8, 10.5936/csbj.201204005.
- Beatrix M. Horváth; Hana Kourová; Szilvia Krisztina Nagy; Edit Németh; Zoltán Magyar; Csaba Papdi; Zaki Ahmad; Gabino F Sanchez‐Perez; Serena Perilli; Ikram Blilou; et al. Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. The EMBO Journal 2017, 36, 1261-1278, 10.15252/embj.201694561.
- Beatrix M. Horváth; Hana Kourová; Szilvia Krisztina Nagy; Edit Németh; Zoltán Magyar; Csaba Papdi; Zaki Ahmad; Gabino F Sanchez‐Perez; Serena Perilli; Ikram Blilou; et al. Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. The EMBO Journal 2017, 36, 1261-1278, 10.15252/embj.201694561.
This entry is offline, you can click here to edit this entry!
