Salinity stress is one of the major threats to agricultural productivity across the globe. Research in the past three decades, therefore, has focused on analyzing the effects of salinity stress on the plants. Evidence gathered over the years supports the role of ethylene as a key regulator of salinity stress tolerance in plants. This gaseous plant hormone regulates many vital cellular processes starting from seed germination to photosynthesis for maintaining the plants’ growth and yield under salinity stress. Ethylene modulates salinity stress responses largely via maintaining the homeostasis of Na+/K+, nutrients, and reactive oxygen species (ROS) by inducing antioxidant defense in addition to elevating the assimilation of nitrates and sulfates. Moreover, a cross-talk of ethylene signaling with other phytohormones has also been observed, which collectively regulate the salinity stress responses in plants.
- ROS
- ethylene
- antioxidants
- salinity stress
- photosynthesis
- programmed cell death
- seed germination
- hormone cross-talk
1. Introduction
2. Ethylene is a Key Modulator of Salinity Stress Responses in Plants
2.1. Salinity Stress and Ethylene Receptors
2.2. Salinity Stress and EIN Proteins
2.3. Effects of Salinity Stress on ERFs and other Ethylene-Responsive Transcription Factors
3. Seed Germination Regulated by Ethylene under Salinity Stress
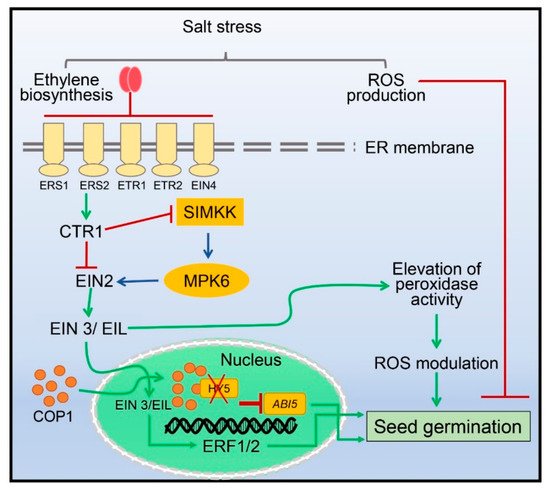
4. Fine-Tuning of Photosynthetic Machinery by Ethylene during Salinity Stress
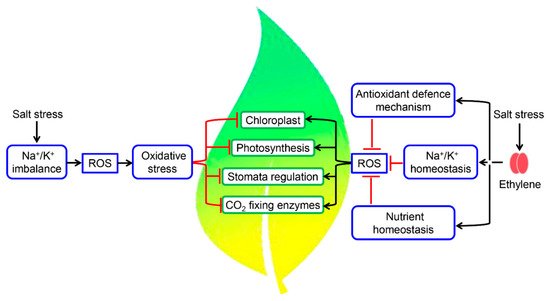
References
- Neljubow, D. Uber die horizontale Nutation der Stengel von Pisum Sativum und einiger anderen Planzen. Bot. Cent. Beih. 1901, 10, 128–139.
- Hua, J. Isolation of Components Involved in Ethylene Signaling. In Ethylene in Plants; Springer: Dordrecht, The Netherlands, 2015; pp. 27–44. ISBN 9401794839.
- Abeles, F.B.; Morgan, P.W.; Salinityveit, M.E., Jr. Ethylene in Plant Biology; Academic Press: San Diego, CA, USA, 2012; ISBN 0080916287.
- Burg, S.P.; Burg, E.A. Molecular requirements for the biological activity of ethylene. Plant Physiol. 1967, 42, 144–152.
- Guzman, P.; Ecker, J.R. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 1990, 2, 513–523.
- Gómez-Cadenas, A.; Tadeo, F.R.; Primo-Millo, E.; Talón, M. Involvement of abscisic acid and ethylene in the responses of citrus seedlings to salinity shock. Physiol. Plant. 1998, 103, 475–484.
- Arraes, F.B.M.; Beneventi, M.A.; de Sa, M.E.L.; Paixao, J.F.R.; Albuquerque, E.V.S.; Marin, S.R.R.; Purgatto, E.; Nepomuceno, A.L.; Grossi-de-Sa, M.F. Implications of ethylene biosynthesis and signaling in soybean drought stress tolerance. BMC Plant Biol. 2015, 15, 213.
- Gupta, R.; Min, C.W.; Kim, S.W.; Yoo, J.S.; Moon, A.R.; Shin, A.Y.; Kwon, S.Y.; Kim, S.T. A TMT-Based Quantitative Proteome Analysis to Elucidate the TSWV Induced Signaling Cascade in Susceptible and Resistant Cultivars of Solanum lycopersicum. Plants 2020, 9, 290.
- Gupta, R.; Min, C.W.; Kim, Y.J.; Kim, S.T. Identification of Msp1-Induced Signaling Components in Rice Leaves by Integrated Proteomic and Phosphoproteomic Analysis. Int. J. Mol. Sci. 2019, 20, 4135.
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681.
- Siddikee, M.A.; Chauhan, P.S.; Sa, T. Regulation of ethylene biosynthesis under salinity stress in red pepper (Capsicum annuum L.) by 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase-producing halotolerant bacteria. J. Plant Growth Regul. 2012, 31, 265–272.
- Freitas, V.S.; de Souza Miranda, R.; Costa, J.H.; de Oliveira, D.F.; de Oliveira Paula, S.; de Castro Miguel, E.; Freire, R.S.; Prisco, J.T.; Gomes-Filho, E. Ethylene triggers salinity tolerance in maize genotypes by modulating polyamine catabolism enzymes associated with H2O2 production. Environ. Exp. Bot. 2018, 145, 75–86.
- Xu, L.; Xiang, G.; Sun, Q.; Ni, Y.; Jin, Z.; Gao, Z.; Yao, Y. Melatonin enhances salinity tolerance by promoting MYB108A-mediated ethylene biosynthesis in grapevines. Hortic. Res. 2019, 6, 114.
- Zhang, M.; Hong, L.Z.; Gu, M.F.; Wu, C.D.; Zhang, G. Transcriptome analyses revealed molecular responses of Cynanchum auriculatum leaves to saline stress. Sci. Rep. 2020, 10, 449.
- Kim, S.W.; Min, C.W.; Gupta, R.; Jo, I.H.; Bang, K.H.; Kim, Y.C.; Kim, K.H.; Kim, S.T. Proteomics analysis of early salinity-responsive proteins in ginseng (Panax ginseng CA Meyer) leaves. Korean J. Med. Crop. Sci. 2014, 22, 398–404.
- Bray, E.A.; Bailey-Serres, J.; Weretilnyk, E. Responses to abiotic stress. In Biochemistry and Molecular Biology of Plants; Buchanan, B., Gruissem, W., Jones, R., Eds.; American Society of Plant Physiology: Rockville, MD, USA, 2000; pp. 1158–1203.
- Pitman, M.G.; Läuchli, A. Global Impact of Salinity and Agricultural Ecosystems. In Salinity: Environment-Plants-Molecules; Springer: Dordrecht, The Netherlands, 2002; pp. 3–20.
- Polle, A.; Chen, S. On the salinityy side of life: Molecular, physiological and anatomical adaptation and acclimation of trees to extreme habitats. Plant Cell Environ. 2015, 38, 1794–1816.
- Ghatak, A.; Chaturvedi, P.; Paul, P.; Agrawal, G.K.; Rakwal, R.; Kim, S.T.; Weckwerth, W.; Gupta, R. Proteomics survey of Solanaceae family: Current status and challenges ahead. J. Proteom. 2017, 169, 41–57.
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salinity stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560.
- Qadir, M.; Quillérou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J.; Drechsel, P.; Noble, A.D. Economics of salinity-induced land degradation and restoration. Nat. Resour. Forum 2014, 38, 282–295.
- Neumann, P.M. Inhibition of root growth by salinity stress: Toxicity or an adaptive biophysical response? In Structure and Function of Roots; Springer: Dordrecht, The Netherlands, 1995; pp. 299–304.
- Rasool, S.; Hameed, A.; Azooz, M.M.; Siddiqi, T.O.; Ahmad, P. Salinity stress: Causes, types and responses of plants. In Ecophysiology and Responses of Plants under Salinity Stress; Springer: New York, NY, USA, 2013; pp. 1–24.
- Muhammad, N.; Samina, B.; Yasmin, K.; Roqayya, M.; Zabta, K.S.; Abdul, L.K.; Ajmal, K.; Ahmed, A. Plant growth promoting bacteria as an alternative strategy for salinity tolerance in plants: A review. Microbiol. Res. 2018, 209, 21–32.
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158.
- Munns, R. Genes and salinity tolerance: Bringing them together. New Phytol. 2005, 167, 645–663.
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salinity-stress responses. New Phytol. 2018, 217, 523–539.
- Fricke, W. Rapid and tissue-specific accumulation of solutes in the growth zone of barley leaves in response to salinity. Planta 2004, 219, 515–525.
- James, R.A.; von Caemmerer, S.; Condon, A.T.; Zwart, A.B.; Munns, R. Genetic variation in tolerance to the osmotic stress component of salinity stress in durum wheat. Funct. Plant Biol. 2008, 35, 111–123.
- Møller, I.M.; Jensen, P.E.; Hansson, A. Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 2007, 58, 459–481.
- Fichman, Y.; Miller, G.; Mittler, R. Whole-plant live imaging of reactive oxygen species. Mol. Plant 2019, 12, 1203–1210.
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19.
- Parida, A.K.; Das, A.B. Salinity tolerance and salinity effect on plants: A review. Ecotoxicol. Environ. Safe 2005, 60, 324–349.
- Younis, M.E.; Hasaneen, M.N.A.; Kazamel, A.M.S. Exogenously applied ascorbic acid ameliorates detrimental effects of NaCl and mannitol stress in Vicia faba seedlings. Protoplasma 2010, 239, 39–48.
- Wang, Y.; Wu, J.; Choi, Y.W.; Jun, T.H.; Kwon, S.W.; Choi, I.S.; Kim, Y.C.; Gupta, R.; Kim, S.T. Expression Analysis of Oryza sativa Ascorbate Peroxidase 1 (OsAPx1) in Response to Different Phytohormones and Pathogens. J. Life Sci. 2015, 25, 1091–1097.
- Wang, Y.; Wu, J.; Kim, S.G.; Tsuda, K.; Gupta, R.; Park, S.Y.; Kim, S.T.; Kang, K.Y. Magnaporthe oryzae-secreted protein MSP1 induces cell death and elicits defense responses in rice. Mol. Plant-Microbe Interact. 2016, 29, 299–312.
- Yang, L.; Zu, Y.G.; Tang, Z.H. Ethylene improves Arabidopsis salinity tolerance mainly via retaining K+ in shoots and roots rather than decreasing tissue Na+ content. Environ. Exp. Bot. 2013, 86, 60–69.
- Gharbi, E.; Martínez, J.P.; Benahmed, H.; Lepoint, G.; Vanpee, B.; Quinet, M.; Lutts, S. Inhibition of ethylene synthesis reduces salinity-tolerance in tomato wild relative species Solanum chilense. J. Plant Physiol. 2017, 210, 24–37.
- Shakar, M.; Yaseen, M.; Mahmood, R.; Ahmad, I. Calcium carbide induced ethylene modulate biochemical profile of Cucumis sativus at seed germination stage to alleviate salinity stress. Sci. Hortic. 2016, 213, 179–185.
- Tavladoraki, P.; Cona, A.; Federico, R.; Tempera, G.; Viceconte, N.; Saccoccio, S.; Battaglia, V.; Toninello, A.; Agostinelli, E. Polyamine catabolism: Target for antiproliferative therapies in animals and stress tolerance strategies in plants. Amino Acids 2012, 42, 411–426.
- Müller, M.; Munnébosch, S. Ethylene response factors. A key regulatory hub in hormone and stress signaling. Plant Physiol. 2015, 169, 32–41.
- Yang, C.; Ma, B.; He, S.J.; Xiong, Q.; Duan, K.X.; Yin, C.C.; Chen, H.; Lu, X.; Chen, S.H.; Zhang, J.H. MAOHUZI6/ETHYLENE INSENSITIVE3-LIKE1 and ETHYLENE INSENSITIVE3-LIKE2 regulate ethylene response of roots and coleoptiles and negatively affect salinity tolerance in rice. Plant Physiol. 2015, 169, 148–165.
- Sajid, H.; Chu, Z.; Zhigang, B.; Xiaochuang, C.; Lianfeng, Z.; Azhar, H.; Zhu, C.; Fahad, S.; James, A.B.; Zhang, J.; et al. Effects of 1-Methylcyclopropene on rice growth characteristics and superior and inferior spikelet development under salinity stress. J. Plant Growth Regul. 2018, 37, 1368–1384.
- Hussain, S.; Bai, Z.; Huang, J.; Cao, X.; Zhu, L.; Zhu, C.; Zhang, J. 1-Methylcyclopropene Modulates Physiological, Biochemical, and Antioxidant Responses of Rice to Different Salinity Stress Levels. Front. Plant Sci. 2019, 10, 124.
- Peng, Z.; He, S.; Gong, W.; Sun, J.; Pan, Z.; Xu, F.; Lu, Y.; Du, X. Comprehensive analysis of differentially expressed genes and transcriptional regulation induced by salt stress in two contrasting cotton genotypes. BMC Genom. 2014, 15, 760.
- Zhao, X.C.; Schaller, G.E. Effect of salinity and osmotic stress upon expression of the ethylene receptor ETR1 in Arabidopsis thaliana. FEBS Lett. 2004, 562, 189–192.
- Zhou, H.L.; Cao, W.H.; Cao, Y.R.; Liu, J.; Hao, Y.J.; Zhang, J.S.; Chen, S.Y. Roles of ethylene receptor NTHK1 domains in plant growth, stress response and protein phosphorylation. FEBS Lett. 2006, 580, 1239–1250.
- Cao, W.H.; Liu, J.; He, X.J.; Mu, R.L.; Zhou, H.L.; Chen, S.Y.; Zhang, J.S. Modulation of ethylene responses affects plant salinity-stress responses. Plant Physiol. 2007, 143, 707–719.
- Wilson, R.L.; Kim, H.; Bakshi, A.; Binder, B.M. The ethylene receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 have contrasting roles in seed germination of Arabidopsis during salinity stress. Plant Physiol. 2014, 165, 1353–1366.
- Cao, W.H.; Liu, J.; Zhou, Q.Y.; Cao, Y.R.; Zheng, S.F.; Du, B.X.; Zang, J.S.; Chen, S.Y. Expression of tobacco ethylene receptor NTHK1 alters plant responses to salinity stress. Plant Cell Environ. 2006, 29, 1210–1219.
- Chen, T.; Liu, J.; Lei, G.; Liu, Y.F.; Li, Z.G.; Tao, J.J.; Hao, Y.J.; Cao, Y.R.; Lin, Q.; Zhang, W.K.; et al. Effects of tobacco Ethylene receptor mutations on receptor kinase activity, plant growth and stress responses. Plant Cell Physiol. 2009, 50, 1636–1650.
- Cao, Y.R.; Chen, H.W.; Li, Z.G.; Tao, J.J.; Ma, B.; Zhang, W.K.; Chen, S.Y.; Zhang, J.S. Tobacco ankyrin protein NEIP2 interacts with ethylene receptor NTHK1 and regulates plant growth and stress responses. Plant Cell Physiol. 2015, 56, 803–818.
- Jiang, C.; Belfield, E.J.; Cao, Y.; Smith, J.A.; Harberd, N.P. An Arabidopsis soil-salinity-tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. Plant Cell. 2013, 25, 3535–3552.
- Achard, P.; Cheng, H.; De Grauwe, L.; Decat, J.; Schoutteten, H.; Moritz, T.; Van Der Straeten, D.; Peng, J.; Harberd, N.P. Integration of plant responses to environmentally activated phytohormonal signals. Science 2006, 311, 91–94.
- Lei, G.; Shen, M.; Li, Z.G.; Zhang, B.; Duan, K.X.; Wang, N.; Cao, Y.R.; Zhang, W.K.; Ma, B.; Ling, H.Q.; et al. EIN2 regulates salinity stress response and interacts with a MA3 domain-containing protein ECIP1 in Arabidopsis. Plant Cell Environ. 2011, 34, 1678–1692.
- Ge, X.M.; Cai, H.L.; Lei, X.; Zhou, X.; Yue, M.; He, J.M. Heterotrimeric G protein mediates ethylene-induced stomatal closure via hydrogen peroxide synthesis in Arabidopsis. Plant J. 2015, 82, 138–150.
- Tao, J.J.; Chen, H.W.; Ma, B.; Zhang, W.K.; Chen, S.Y.; Zhang, J.S. The role of ethylene in plants under salinity stress. Front. Plant Sci. 2015, 6, 1059.
- Zhang, L.X.; Li, Z.F.; Quan, R.D.; Li, G.J.; Wang, R.G.; Huang, R.F. An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salinity response in Arabidopsis. Plant Physiol. 2011, 157, 854–865.
- Quan, R.; Wang, J.; Yang, D.; Zhang, H.; Zhang, Z.; Huang, R. EIN3 and SOS2 synergistically modulate plant salinity tolerance. Sci. Rep. 2017, 7, 44637–44647.
- Qin, H.; Wang, J.; Chen, X.; Wang, F.; Peng, P.; Zhou, Y.; Miao, Y.; Zhang, Y.; Gao, Y.; Qi, Y.; et al. Rice Os DOF 15 contributes to ethylene-inhibited primary root elongation under salinity stress. New Phytol. 2019, 223, 798–813.
- Jin, J.; Duan, J.; Shan, C.; Mei, Z.; Chen, H.; Feng, H.; Zhu, J.; Cai, W. Ethylene insensitive3-like2 (OsEIL2) confers stress sensitivity by regulating OsBURP16, the β subunit of polygalacturonase (PG1β-like) subfamily gene in rice. Plant Sci. 2020, 292, 110353.
- Liu, C.; Li, J.; Zhu, P.; Yu, J.; Hou, J.; Wang, C.; Long, D.; Yu, M.; Zhao, A. Mulberry EIL3 confers salinity and drought tolerances and modulates ethylene biosynthetic gene expression. PeerJ 2019, 7, e6391.
- Wang, C.; Li, J.; Yuan, M. Salinity tolerance requires cortical microtubule reorganization in Arabidopsis. Plant Cell Physiol. 2007, 48, 1534–1547.
- Zhang, Q.; Lin, F.; Mao, T.; Nie, J.; Yan, M.; Yuan, M.; Zhang, W. Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salinity stress in Arabidopsis. Plant Cell 2012, 24, 4555–4576.
- Le, J.; Vandenbusscheb, F.; Straetenb, D.V.D.; Verbelen, J.P. Position and cell 786 type-dependent microtubule reorientation characterizes the early response of the 787 Arabidopsis root epidermis to ethylene. Physiol. Plant. 2004, 121, 513–519.
- Verbelen, J.P.; Jie, L.; Vissenberg, K.; Cnodder, T.D.; Vandenbussche, F.; Sugimoto, K.; Straeten, D.V.D. Microtubules and the control of cell elongation in Arabidopsis roots. In The Plant Cytoskeleton: A Key Tool for Agro-Biotechnology; Springer: Dordrecht, The Netherlands, 2008; pp. 73–90.
- Sun, J.; Ma, Q.; Mao, T. Ethylene regulates the Arabidopsis microtubule-associated protein WAVE-DAMPENED2-LIKE5 in etiolated hypocotyl elongation. Plant Physiol. 2015, 169, 325–337.
- Ma, Q.; Sun, J.; Mao, T. Microtubule bundling plays a role in ethylene-mediated cortical microtubule reorientation in etiolated Arabidopsis hypocotyls. J. Cell Sci. 2016, 129, 2043–2051.
- Dou, L.; He, K.; Higaki, T.; Wang, X.; Mao, T. Ethylene signaling modulates cortical microtubule reassembly in response to salinity stress. Plant Physiol. 2018, 176, 2071–2081.
- Cheng, M.C.; Liao, P.M.; Kuo, W.W.; Lin, T.P. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013, 162, 1566–1582.
- Li, Y.; Zhang, H.; Zhang, Q.; Liu, Q.; Zhai, H.; Zhao, N.; He, S. An AP2/ERF gene, IbRAP2-12, from sweetpotato is involved in salinity and drought tolerance in transgenic Arabidopsis. J. Plant Sci. 2019, 281, 19–30.
- Djemal, R.; Khoudi, H. TdSHN1, a WIN1/SHN1-type transcription factor, imparts multiple abiotic stress tolerance in transgenic tobacco. Environ. Exp. Bot. 2016, 131, 89–100.
- An, J.P.; Zhang, X.W.; Xu, R.R.; You, C.X.; Wang, X.F.; Hao, Y.J. Apple MdERF4 negatively regulates salinity tolerance by inhibiting MdERF3 transcription. Plant Sci. 2018, 276, 181–188.
- Cheng, Z.; Zhang, X.; Zhao, K.; Yao, W.; Li, R.; Zhou, B.; Jiang, T. Over-expression of ERF38 gene enhances salt and osmotic tolerance in transgenic poplar. Front. Plant Sci. 2019, 10, 1375.
- Yang, S.F.; Hoffman, N.E. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 1984, 35, 155–189.
- Wang, K.; Li, H.; Ecker, J.R. Ethylene biosynthesis and signaling networks. Plant Cell 2002, 14, 131–151.
- Sauter, M.; Lorbiecke, R.; OuYang, B.; Pochapsky, T.C.; Rzewuski, G. The immediate-early ethylene response gene OsARD1 encodes an acireductone dioxygenase involved in recycling of the ethylene precursor S-adenosylmethionine. Plant J. 2005, 44, 718–729.
- Liang, S.; Xiong, W.; Yin, C.; Xie, X.; Jin, Y.J.; Zhang, S.; Yang, B.; Ye, G.; Chen, S.; Luan, W.J. Overexpression of OsARD1 Improves Submergence, Drought, and Salinity Tolerances of Seedling Through the Enhancement of Ethylene Synthesis in Rice. Front. Plant Sci. 2019, 10, 1088.
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523.
- Holdsworth, M.J.; Bentsink, L.; Soppe, W.J. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008, 179, 33–54.
- Weitbrecht, K.; Müller, K.; Leubner-Metzger, G. First off the mark: Early seed germination. J. Exp. Bot. 2011, 62, 3289–3309.
- Ghassemian, M.; Nambara, E.; Cutler, S.; Kawaide, H.; Kamiya, Y.; McCourt, P. Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 2000, 12, 1117–1126.
- Cao, Y.R.; Chen, S.Y.; Zhang, J.S. Ethylene signaling regulates salinity stress response: An overview. Plant Signal. Behav. 2008, 3, 761–763.
- Yu, Y.; Wang, J.; Shi, H.; Gu, J.; Dong, J.; Deng, X.W.; Huang, R. Salinity stress and ethylene antagonistically regulate nucleocytoplasmic partitioning of COP1 to control seed germination. Plant Physiol. 2016, 170, 2340–2350.
- Lin, Y.; Yang, L.; Paul, M.; Zu, Y.; Tang, Z. Ethylene promotes germination of Arabidopsis seed under salinity by decreasing reactive oxygen species: Evidence for the involvement of nitric oxide simulated by sodium nitroprusside. Plant Physiol. Biochem. 2013, 73, 211–218.
- Chang, C.; Wang, B.; Shi, L.; Li, Y.; Duo, L.; Zhang, W. Alleviation of salinity stress-induced inhibition of seed germination in cucumber (Cucumis sativus L.) by ethylene and glutamate. J. Plant Physiol. 2010, 167, 1152–1156.
- Zhang, J.Y.; Luo, H.T.; Guo, Z.R. Overexpression of Malus hupehensis MhSHN1 Gene Enhances Salinity and Osmotic Stress Tolerance in Transgenic Tobacco Plants. Russ. J. Plant Physiol. 2018, 65, 857–864.
- Lovato, M.B.; de Lemos Filho, J.P.; Martins, P.S. Growth responses of Stylosanthes humilis (Fabaceae) populations to saline stress. Environ. Exp. Bot. 1999, 41, 145–153.
- Silva, P.O.; Medina, E.F.; Barros, R.S.; Ribeiro, D.M. Germination of salt-stressed seeds as related to the ethylene biosynthesis ability in three Stylosanthes species. J. Plant Physiol. 2014, 171, 14–22.
- Silva, N.C.; de Souza, G.A.; Pimenta, T.M.; Brito, F.A.; Picoli, E.A.; Zsögön, A.; Ribeiro, D.M. Salinity stress inhibits germination of Stylosanthes humilis seeds through abscisic acid accumulation and associated changes in ethylene production. Plant Physiol. Biochem. 2018, 130, 399–407.
- Zhang, S.; Yang, R.; Huo, Y.; Liu, S.; Yang, G.; Huang, J.; Zheng, C.; Wu, C. Expression of cotton PLATZ1 in transgenic Arabidopsis reduces sensitivity to osmotic and salinity stress for germination and seedling establishment associated with modification of the abscisic acid, gibberellin, and ethylene signalling pathways. BMC Plant Biol. 2018, 18, 1–11.
- Ma, L.; Hu, L.; Fan, J.; Amombo, E.; Khaldun, A.B.M.; Zheng, Y.; Chen, L. Cotton GhERF38 gene is involved in plant response to salinity/drought and ABA. Ecotoxicology 2017, 26, 841–854.
- Safari, D.; Jamali, F.; Nooryazdan, H.R.; Bayat, F. Evaluation of ACC deaminase producing ‘Pseudomonas fluorescens’ strains for their effects on seed germination and early growth of wheat under salinity stress. Aust. J. Crop. Sci. 2018, 12, 413.
- Li, H.; Lei, P.; Pang, X.; Li, S.; Xu, H.; Xu, Z.; Feng, X. Enhanced tolerance to salinity stress in canola (Brassica napus L.) seedlings inoculated with the halotolerant Enterobacter cloacae HSNJ4. Appl. Soil Ecol. 2017, 119, 26–34.
- Wang, X.; Hou, C.; Zheng, K.; Li, Q.; Chen, S.; Wang, S. Overexpression of ERF96, a small ethylene response factor gene, enhances salinity tolerance in Arabidopsis. Biol. Plant 2017, 61, 693–701.
- Wu, D.; Ji, J.; Wang, G.; Guan, C.; Jin, C. LchERF, a novel ethylene-responsive 76transcription factor from Lycium chinense, confers salinity tolerance in transgenic tobacco. Plant Cell Rep. 2014, 33, 2033–2045.
- Yang, F.; Chen, H.; Liu, C.; Li, L.; Liu, L.; Han, X.; Wan, Z.; Sha, A. Transcriptome profile analysis of two Vicia faba cultivars with contrasting salinity tolerance during seed germination. Sci. Rep. 2020, 10, 7250.
- Ahmed, W.; Imran, M.; Yaseen, M.; ul Haq, T.; Jamshaid, M.U.; Rukh, S.; Ikram, R.M.; Ali, M.; Ali, A.; Maqbool, M.; et al. Role of salicylic acid in regulating ethylene and physiological characteristics for alleviating salinity stress on germination, growth and yield of sweet pepper. PeerJ 2020, 8, e8475.
- Khan, M.I.R.; Asgher, M.; Khan, N.A. Alleviation of salinity-induced photosynthesis and growth inhibition by salicylic acid involves glycinebetaine and ethylene in mungbean (Vigna radiata L.). Plant Physiol. Biochem. 2014, 80, 67–74.
- Ahanger, M.A.; Agarwal, R.M. Salinity stress induced alterations in antioxidant metabolism and nitrogen assimilation in wheat (Triticum aestivum L.) as influenced by potassium supplementation. Plant Physiol. Biochem. 2017, 115, 449–460.
- Ahanger, M.A.; Qin, C.; Begum, N.; Maodong, Q.; Dong, X.X.; El-Esawi, M.; El-Sheikh, M.A.; Alatar, A.A.; Zhang, L. Nitrogen availability prevents oxidative effects of salinity on wheat growth and photosynthesis by up-regulating the antioxidants and osmolytes metabolism, and secondary metabolite accumulation. BMC Plant Biol. 2019, 19, 1–12.
- Cheeseman, J.M. The integration of activity in saline environments: Problems and perspectives. Funct. Plant Biol. 2013, 40, 759–774.
- Bose, J.; Munns, R.; Shabala, S.; Gilliham, M.; Pogson, B.; Tyerman, S.D. Chloroplast function and ion regulation in plants growing on saline soils: Lessons from halophytes. J. Exp. Bot. 2017, 68, 3129–3143.
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salinity tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433.
- Liu, C.; Zhao, X.; Yan, J.; Yuan, Z.; Gu, M. Effects of Salinity Stress on Growth, Photosynthesis, and Mineral Nutrients of 18 Pomegranate (Punica granatum) Cultivars. Agronomy 2020, 10, 27.
- Brito, F.A.; Pimenta, T.M.; Henschel, J.M.; Martins, S.C.; Zsögön, A.; Ribeiro, D.M. Elevated CO2 improves assimilation rate and growth of tomato plants under progressively higher soil salinity by decreasing abscisic acid and ethylene levels. Environ. Exp. Bot. 2020, 176, 104050.
- Murata, N.; Takahashi, S.; Nishiyama, Y.; Allakhverdiev, S.I. Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta 2007, 1767, 414–421.
- Liu, C.; Li, L.L.; Li, G.Z.; Hao, L. Ethylene insensitive mutation improves Arabidopsis plant tolerance to NO2 exposure. Ecotoxicol. Environ. Saf. 2020, 189, 110043.
- Sarafi, E.; Chatzissavvidis, C.; Therios, I. Effect of calcium and boron on the ion status, carbohydrate and proline content, gas exchange parameters and growth performance of pomegranate cv. Wonderful plants grown under NaCl stress. Türk Tarım ve Doğa Bilimleri Dergisi 2014, 1, 1606–1617.
- Sun, Y.; Niu, G.; Masabni, J.G.; Ganjegunte, G. Relative salinity tolerance of 22 pomegranate (Punica granatum) cultivars. HortScience 2018, 53, 1513–1519.
- Pan, Y.J.; Liu, L.; Lin, Y.C.; Zu, Y.G.; Li, L.P.; Tang, Z.H. Ethylene antagonizes salinity-induced growth retardation and cell death process via transcriptional controlling of ethylene-, BAG-and senescence-associated genes in Arabidopsis. Front. Plant Sci. 2016, 7, 696.
- Splivallo, R.; Fischer, U.; Göbel, C.; Feussner, I.; Karlovsky, P. Truffles regulate plant root morphogenesis via the production of auxin and ethylene. Plant Physiol. 2009, 150, 2018–2029.
- Nouri, M.Z.; Moumeni, A.; Komatsu, S. Abiotic stresses: Insight into gene regulation and protein expression in photosynthetic pathways of plants. Int. J. Mol. Sci. 2015, 16, 20392–20416.
- De Zélicourt, A.; Synek, L.; Saad, M.M.; Alzubaidy, H.; Jalal, R.; Xie, Y.; Andres-Barrao, C.; Rolli, E.; Guerard, F.; Mariappan, K.G.; et al. Ethylene induced plant stress tolerance by Enterobacter sp. SA187 is mediated by 2-keto-4-methylthiobutyric acid production. PLoS Genet. 2018, 14, e1007273.
- Ji, J.; Yuan, D.; Jin, C.; Wang, G.; Li, X.; Guan, C. Enhancement of growth and salinity tolerance of rice seedlings (Oryza sativa L.) by regulating ethylene production with a novel halotolerant PGPR strain Glutamicibacter sp. YD01 containing ACC deaminase activity. Acta Physiol. Plant. 2020, 42, 1–17.
- Foyer, C.H.; Shigeoka, S. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 2011, 155, 93–100.
- Considine, M.J.; Foyer, C.H. Redox regulation of plant development. Antioxid. Redox Signal. 2014, 21, 1305–1326.
- Noctor, G.; Mhamdi, A.; Foyer, C.H. The roles of reactive oxygen metabolism in drought: Not so cut and dried. Plant Physiol. 2014, 164, 1636–1648.
- Iqbal, N.; Umar, S.; Khan, N.A. Nitrogen availability regulates proline and ethylene production and alleviates salinity stress in mustard (Brassica juncea). J. Plant Physiol. 2015, 178, 84–91.
- Zhang, F.; Wan, X.; Zhong, Y. Nitrogen as an important detoxification factor to cadmium stress in poplar plants. J. Plant Interact. 2014, 9, 249–258.
