Lysophosphatidic acid receptor 3 (LPA3) is implicated in different physiological and pathological functions through activation of different signal pathways, the result of the regulation process of this receptor. The knowledge of regulating LPA3 could be a crucial element for defined their roles in health and disease.
- lysophosphatidic acid 3 receptor
- receptor phosphorylation
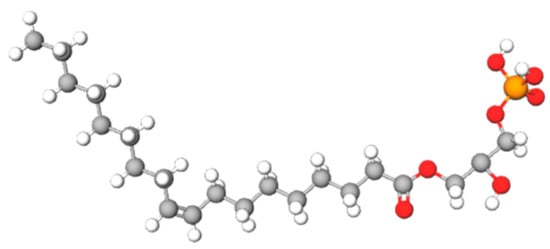
- lysophosphatidic acid
- PKC
- GRK
1. Introducción
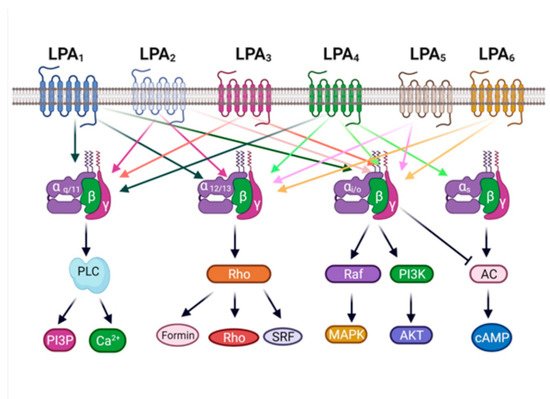
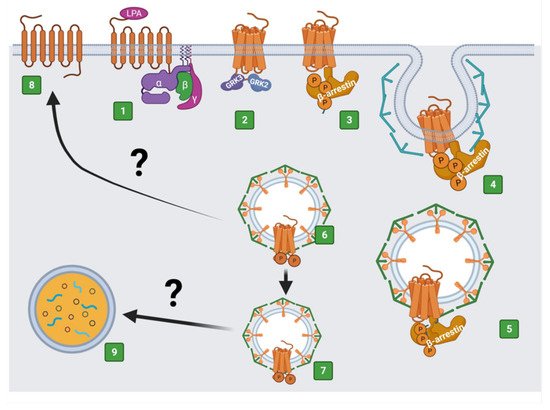
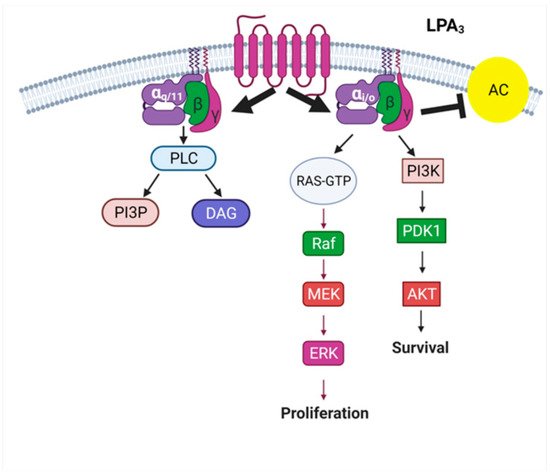
2. The LPA3 Receptor: Structure and Function
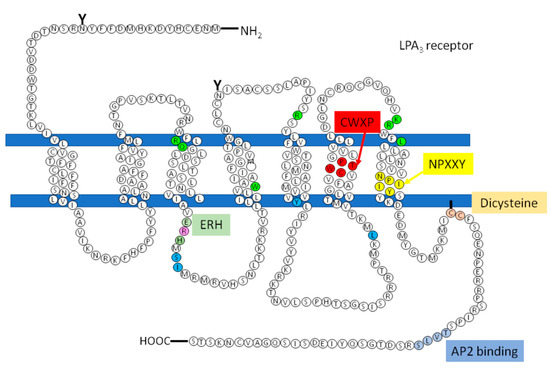
However, there are receptors of the same family that do not present this sequence that could be involved in the recruitment of the G protein, how is the LPA3, so according to studies carried out by Zhou and coworkers, in which it is proposed that in response to agonist-induced conformational changes, residues in transmembrane domains 3, 5, and 6 interact with and activate G proteins[77]. These residues were found in the structure of the LPA3 receptor as shown in Figure 5 (indicated in cerulean).
| Subfamilies | GRKs | Domains of Interest |
|---|---|---|
| Visual GRKs | GRK1 and GRK7 | Prenylation |
| GRK2 or βARK GRK4 |
GRK2 and GRK3 GRK4, GRK5 and GRK6 |
Pleckstrin homology Palmitoylation, polybasic hydrophobic domains |
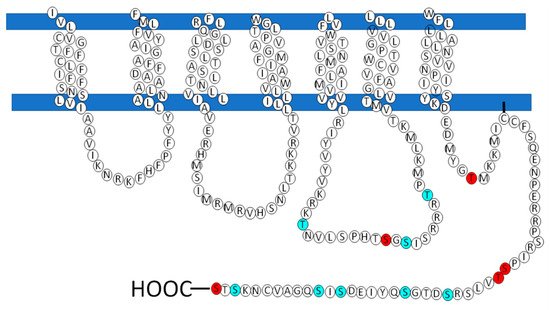
| Posición | Aminoácidos | PKC / PKA |
|---|---|---|
| 130 | S | PKA |
| 217 | T | PKCα / PKCδ / PKCγ |
| 233 | T | PKA / PKCδ / PKCι / PKCζ |
| 243 | T | PKCi / PKCζ |
| 321 | S | PKA / PKCδ / |
| 325 | S | PKA / PKC / PKCε |
| 341 | S | PKCε |
| 351 | S | PKCε |
This entry is adapted from the peer-reviewed paper 10.3390/ijms22136704
References
- Yasuyuki Kihara; Michael Maceyka; Sarah Spiegel; Jerold Chun; Lysophospholipid receptor nomenclature review: IUPHAR Review 8. Journal of Cerebral Blood Flow & Metabolism 2014, 171, 3575-3594, 10.1111/bph.12678.
- Fang Yang; Guo-Xun Chen; Production of extracellular lysophosphatidic acid in the regulation of adipocyte functions and liver fibrosis.. World Journal of Gastroenterology 2018, 24, 4132-4151, 10.3748/wjg.v24.i36.4132.
- Frédérique Gaits; Olivier Fourcade; François Le Balle; Geneviève Gueguen; Bernadette Gaigé; Ama Gassama-Diagne; Josette Fauvel; Jean-Pierre Salles; Gérard Mauco; Marie-Françoise Simon; et al. Lysophosphatidic acid as a phospholipid mediator: pathways of synthesis.. FEBS Letters 1997, 410, 54-58, 10.1016/s0014-5793(97)00411-0.
- Keita Nakanaga; Kotaro Hama; Junken Aoki; Autotaxin--an LPA producing enzyme with diverse functions. The Journal of Biochemistry 2010, 148, 13-24, 10.1093/jb/mvq052.
- Yun C. Yung; Nicole C. Stoddard; Jerold Chun; LPA receptor signaling: pharmacology, physiology, and pathophysiology. Journal of Lipid Research 2014, 55, 1192-1214, 10.1194/jlr.r046458.
- Junken Aoki; Mechanisms of lysophosphatidic acid production. Seminars in Cell & Developmental Biology 2004, 15, 477-489, 10.1016/j.semcdb.2004.05.001.
- Sindhu Ramesh; Manoj Govindarajulu; Vishnu Suppiramaniam; Timothy Moore; Muralikrishnan Dhanasekaran; Autotaxin–Lysophosphatidic Acid Signaling in Alzheimer’s Disease. International Journal of Molecular Sciences 2018, 19, 1827, 10.3390/ijms19071827.
- Ji Woong Choi; Deron R. Herr; Kyoko Noguchi; Yun C. Yung; Chang-Wook Lee; Tetsuji Mutoh; Mu-En Lin; Siew T. Teo; Kristine E. Park; Alycia N. Mosley; et al. LPA Receptors: Subtypes and Biological Actions. Annual Review of Pharmacology and Toxicology 2010, 50, 157-186, 10.1146/annurev.pharmtox.010909.105753.
- Jerold Chun; Timothy Hla; Kevin R. Lynch; Sarah Spiegel; Wouter H. Moolenaar; International Union of Basic and Clinical Pharmacology. LXXVIII. Lysophospholipid Receptor Nomenclature: TABLE 1. Pharmacological Reviews 2010, 62, 579-587, 10.1124/pr.110.003111.
- Daisuke Yasuda; Daiki Kobayashi; Noriyuki Akahoshi; Takayo Ohto-Nakanishi; Kazuaki Yoshioka; Yoh Takuwa; Seiya Mizuno; Satoru Takahashi; Satoshi Ishii; Lysophosphatidic acid–induced YAP/TAZ activation promotes developmental angiogenesis by repressing Notch ligand Dll4. Journal of Clinical Investigation 2019, 129, 4332-4349, 10.1172/jci121955.
- Carol M. Rivera-Lopez; Amy L. Tucker; Kevin R. Lynch; Lysophosphatidic acid (LPA) and angiogenesis. Angiogenesis 2008, 11, 301-310, 10.1007/s10456-008-9113-5.
- Isabel Gross; Anja U. Bräuer; Modulation of lysophosphatidic acid (LPA) receptor activity: the key to successful neural regeneration?. Neural Regeneration Research 2019, 15, 53-54, 10.4103/1673-5374.264452.
- C. Laura Sayas; M. Teresa Moreno-Flores; Jesús Avila; Francisco Wandosell; The Neurite Retraction Induced by Lysophosphatidic Acid Increases Alzheimer's Disease-like Tau Phosphorylation. Journal of Biological Chemistry 1999, 274, 37046-37052, 10.1074/jbc.274.52.37046.
- Jong Hee Choi; Minhee Jang; Yeeun Jang; Ik Hyun Cho; Gintonin, a ginseng-derived ingredient, as a novel therapeutic strategy for Huntington's disease: Activation of the Nrf2 pathway through lysophosphatidic acid receptors. IBRO Reports 2019, 6, S101, 10.1016/j.ibror.2019.07.327.
- Eriko Tanabe; Misaho Kitayoshi; Kyohei Yoshikawa; Ayano Shibata; Kanya Honoki; Nobuyuki Fukushima; Toshifumi Tsujiuchi; Loss of lysophosphatidic acid receptor-3 suppresses cell migration activity of human sarcoma cells. Journal of Receptors and Signal Transduction 2012, 32, 328-334, 10.3109/10799893.2012.738689.
- Kyoko Okabe; Kohei Kato; Miki Teranishi; Mai Okumura; Rie Fukui; Toshio Mori; Nobuyuki Fukushima; Toshifumi Tsujiuchi; Induction of lysophosphatidic acid receptor-3 by 12-O-tetradecanoylphorbol-13-acetate stimulates cell migration of rat liver cells. Cancer Letters 2011, 309, 236-242, 10.1016/j.canlet.2011.06.020.
- Shinya Shano; Kazuki Hatanaka; Shinsuke Ninose; Ryutaro Moriyama; Toshifumi Tsujiuchi; Nobuyuki Fukushima; A lysophosphatidic acid receptor lacking the PDZ-binding domain is constitutively active and stimulates cell proliferation. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 2008, 1783, 748-759, 10.1016/j.bbamcr.2007.11.013.
- Mai Hayashi; Kyoko Okabe; Kohei Kato; Mai Okumura; Rie Fukui; Nobuyuki Fukushima; Toshifumi Tsujiuchi; Differential function of lysophosphatidic acid receptors in cell proliferation and migration of neuroblastoma cells. Cancer Letters 2012, 316, 91-96, 10.1016/j.canlet.2011.10.030.
- Zachariah G. Goldsmith; Ji Hee Ha; Muralidharan Jayaraman; Danny N. Dhanasekaran; Lysophosphatidic Acid Stimulates the Proliferation of Ovarian Cancer Cells via the gep Proto-Oncogene G 12. Genes & Cancer 2011, 2, 563-575, 10.1177/1947601911419362.
- Brigitte Anliker; Ji Woong Choi; Mu-En Lin; Shannon E. Gardell; Richard R. Rivera; Grace Kennedy; Jerold Chun; Lysophosphatidic acid (LPA) and its receptor, LPA1, influence embryonic schwann cell migration, myelination, and cell-to-axon segregation. Glia 2013, 61, 2009-2022, 10.1002/glia.22572.
- Laura Sánchez-Marín; David Ladrón de Guevara-Miranda; M. Carmen Mañas-Padilla; Francisco Alén; Román D. Moreno-Fernández; Caridad Díaz-Navarro; José Pérez-Del Palacio; María García-Fernández; Carmen Pedraza; Francisco J. Pavón; et al. Systemic blockade of LPA1/3 lysophosphatidic acid receptors by ki16425 modulates the effects of ethanol on the brain and behavior. Neuropharmacology 2018, 133, 189-201, 10.1016/j.neuropharm.2018.01.033.
- Yun C. Yung; Nicole C. Stoddard; Hope Mirendil; Jerold Chun; Lysophosphatidic Acid Signaling in the Nervous System. Neuron 2015, 86, 341, 10.1016/j.neuron.2015.03.043.
- Hiroshi Ueda; Hiroyuki Neyama; Keita Sasaki; Chiho Miyama; Ryusei Iwamoto; Lysophosphatidic acid LPA1 and LPA3 receptors play roles in the maintenance of late tissue plasminogen activator-induced central poststroke pain in mice. Neurobiology of Pain 2018, 5, 100020, 10.1016/j.ynpai.2018.07.001.
- Ken Kuwajima; Masahiko Sumitani; Makoto Kurano; Kuniyuki Kano; Masako Nishikawa; Baasanjav Uranbileg; Rikuhei Tsuchida; Toru Ogata; Junken Aoki; Yutaka Yatomi; et al. Lysophosphatidic acid is associated with neuropathic pain intensity in humans: An exploratory study. PLOS ONE 2018, 13, e0207310, 10.1371/journal.pone.0207310.
- Susann Fayyaz; Lukasz Japtok; Fabian Schumacher; Dominik Wigger; Tim Julius Schulz; Kathrin Haubold; Erich Gulbins; Heinz Völler; Burkhard Kleuser; Lysophosphatidic Acid Inhibits Insulin Signaling in Primary Rat Hepatocytes via the LPA3 Receptor Subtype and is Increased in Obesity.. Cellular Physiology and Biochemistry 2017, 43, 445-456, 10.1159/000480470.
- Gábor Tigyi; Mélanie A. Dacheux; Kuan-Hung Lin; Junming Yue; Derek Norman; Zoltán Benyó; Sue Chin Lee; Anti-cancer strategies targeting the autotaxin-lysophosphatidic acid receptor axis: is there a path forward?. Cancer and Metastasis Reviews 2021, 40, 3-5, 10.1007/s10555-021-09955-5.
- Kai Sun; Hui Cai; Xiaoyi Duan; Ya Yang; Min Li; Jingkun Qu; Xu Zhang; Jiansheng Wang; Aberrant expression and potential therapeutic target of lysophosphatidic acid receptor 3 in triple-negative breast cancers. Clinical and Experimental Medicine 2014, 15, 371-380, 10.1007/s10238-014-0306-5.
- Ming Quan; Jiu-Jie Cui; Xiao Feng; Qian Huang; The critical role and potential target of the autotaxin/lysophosphatidate axis in pancreatic cancer. Tumor Biology 2017, 39, 1-11, 10.1177/1010428317694544.
- Jiang Chen; Hongyu Li; Wenda Xu; Xiaozhong Guo; Evaluation of serum ATX and LPA as potential diagnostic biomarkers in patients with pancreatic cancer. BMC Gastroenterology 2021, 21, 1-10, 10.1186/s12876-021-01635-6.
- Jong Han Lee; Donghee Kim; Yoon Sin Oh; Hee-Sook Jun; Lysophosphatidic Acid Signaling in Diabetic Nephropathy.. International Journal of Molecular Sciences 2019, 20, 2850, 10.3390/ijms20112850.
- G Tigyi; R Miledi; Lysophosphatidates bound to serum albumin activate membrane currents in Xenopus oocytes and neurite retraction in PC12 pheochromocytoma cells.. Journal of Biological Chemistry 1992, 267, 21360-21367, 10.1016/s0021-9258(19)36618-9.
- Elisabeth Panther; Marco Idzko; Silvia Corinti; Davide Ferrari; Yared Herouy; Maja Mockenhaupt; Stefan Dichmann; Peter Gebicke-Haerter; Francesco Di Virgilio; Giampiero Girolomoni; et al. The Influence of Lysophosphatidic Acid on the Functions of Human Dendritic Cells. The Journal of Immunology 2002, 169, 4129-4135, 10.4049/jimmunol.169.8.4129.
- Jeremy D. Ward; Danny N. Dhanasekaran; LPA Stimulates the Phosphorylation of p130Cas via G i2 in Ovarian Cancer Cells. Genes & Cancer 2012, 3, 578-591, 10.1177/1947601913475360.
- Jinjing Yang; Jiyao Xu; Xuebin Han; Hao Wang; Yuean Zhang; Jin Dong; Yongzhi Deng; Jingping Wang; Lysophosphatidic Acid Is Associated With Cardiac Dysfunction and Hypertrophy by Suppressing Autophagy via the LPA3/AKT/mTOR Pathway. Frontiers in Physiology 2018, 9, 1315, 10.3389/fphys.2018.01315.
- Yan Liao; Ganggang Mu; Lingli Zhang; Wei Zhou; Jun Zhang; Honggang Yu; Lysophosphatidic Acid Stimulates Activation of Focal Adhesion Kinase and Paxillin and Promotes Cell Motility, via LPA1–3, in Human Pancreatic Cancer. Digestive Diseases and Sciences 2013, 58, 3524-3533, 10.1007/s10620-013-2878-4.
- Junken Aoki; Two pathways for lysophosphatidic acid production. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 2008, 1781, 513-518, 10.1016/j.bbalip.2008.06.005.
- Helgi B. Schiöth; Robert Fredriksson; The GRAFS classification system of G-protein coupled receptors in comparative perspective. General and Comparative Endocrinology 2005, 142, 94-101, 10.1016/j.ygcen.2004.12.018.
- Matthew N. Davies; Andrew Secker; Alex Freitas; Miguel Mendao; Jonathan Ian Timmis; Darren R. Flower; On the hierarchical classification of G protein-coupled receptors. Bioinformatics 2007, 23, 3113-3118, 10.1093/bioinformatics/btm506.
- Yoh Takuwa; Noriko Takuwa; Naotoshi Sugimoto; The Edg Family G Protein-Coupled Receptors for Lysophospholipids: Their Signaling Properties and Biological Activities. The Journal of Biochemistry 2002, 131, 767-771, 10.1093/oxfordjournals.jbchem.a003163.
- Kim A Neve , Jeremy K Seamans, Heather Trantham-Davidson; Dopamine receptor signaling. J Recept Signal Transduct Res 2004, 24, 165-205, 10.1081/rrs-200029981.
- Mu-En Lin; Deron R. Herr; Jerold Chun; Lysophosphatidic acid (LPA) receptors: Signaling properties and disease relevance. Prostaglandins & Other Lipid Mediators 2010, 91, 130-138, 10.1016/j.prostaglandins.2009.02.002.
- Stéphane Dalle; Takeshi Imamura; David W. Rose; Dorothy Sears Worrall; Satoshi Ugi; Christopher J. Hupfeld; Jerrold M. Olefsky; Insulin Induces Heterologous Desensitization of G Protein-Coupled Receptor and Insulin-Like Growth Factor I Signaling by Downregulating β-Arrestin-1. Molecular and Cellular Biology 2002, 22, 6272-6285, 10.1128/mcb.22.17.6272-6285.2002.
- Davide Calebiro; Amod Godbole; Internalization of G-protein-coupled receptors: Implication in receptor function, physiology and diseases. Best Practice & Research Clinical Endocrinology & Metabolism 2018, 32, 83-91, 10.1016/j.beem.2018.01.004.
- Qisheng Liu; Mark S. Bee; Agnes Schonbrunn; Site Specificity of Agonist and Second Messenger-Activated Kinases for Somatostatin Receptor Subtype 2A (Sst2A) Phosphorylation. Molecular Pharmacology 2009, 76, 68-80, 10.1124/mol.108.054262.
- Richard Premont; James Inglese; Robert J. Lefkowitz; Protein kinases that phosphorylate activated G protein‐coupled receptors. The FASEB Journal 1995, 9, 175-182, 10.1096/fasebj.9.2.7781920.
- Marco A. Alfonzo-Méndez; Gabriel Carmona-Rosas; David A. Hernández-Espinosa; M. Teresa Romero-Ávila; J. Adolfo García-Sáinz; Different phosphorylation patterns regulate α1D-adrenoceptor signaling and desensitization. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 2018, 1865, 842-854, 10.1016/j.bbamcr.2018.03.006.
- Denise Wootten; Arthur Christopoulos; Maria Marti-Solano; M. Madan Babu; Patrick M. Sexton; Mechanisms of signalling and biased agonism in G protein-coupled receptors. Nature Reviews Molecular Cell Biology 2018, 19, 638-653, 10.1038/s41580-018-0049-3.
- Stuart J. Mundell; Matthew L. Jones; Adam R. Hardy; Johanna F. Barton; Stephanie Beaucourt; Pamela B. Conley; Alastair W. Poole; Distinct Roles for Protein Kinase C Isoforms in Regulating Platelet Purinergic Receptor Function. Molecular Pharmacology 2006, 70, 1132-1142, 10.1124/mol.106.023549.
- Mandi M. Murph; Launa A. Scaccia; Laura A. Volpicelli; Harish Radhakrishna; Agonist-induced endocytosis of lysophosphatidic acid-coupled LPA1/EDG-2 receptors via a dynamin2- and Rab5-dependent pathway. Journal of Cell Science 2003, 116, 1969-1980, 10.1242/jcs.00397.
- Ana C Magalhaes; Henry A. Dunn; Stephen Sg Ferguson; Regulation of GPCR activity, trafficking and localization by GPCR-interacting proteins. Journal of Cerebral Blood Flow & Metabolism 2012, 165, 1717-1736, 10.1111/j.1476-5381.2011.01552.x.
- Fangtian Huang; Anastasia Khvorova; William Marshall; Alexander Sorkin; Analysis of Clathrin-mediated Endocytosis of Epidermal Growth Factor Receptor by RNA Interference. Journal of Biological Chemistry 2004, 279, 16657-16661, 10.1074/jbc.c400046200.
- Braden T. Lobingier; Mark Von Zastrow; When trafficking and signaling mix: How subcellular location shapes G protein-coupled receptor activation of heterotrimeric G proteins. Traffic 2018, 20, 130-136, 10.1111/tra.12634.
- Stephan K. Böhm; Lev M. Khitin; Steven P. Smeekens; Eileen F. Grady; Donald G. Payan; Nigel W. Bunnett; Identification of Potential Tyrosine-containing Endocytic Motifs in the Carboxyl-tail and Seventh Transmembrane Domain of the Neurokinin 1 Receptor. Journal of Biological Chemistry 1996, 272, 2363-2372, 10.1074/jbc.272.4.2363.
- Alexander Zaslavsky; Lisam Shanjukumar Singh; Haiyan Tan; Huawen Ding; Zicai Liang; Yan Xu; Homo- and hetero-dimerization of LPA/S1P receptors, OGR1 and GPR4. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 2006, 1761, 1200-1212, 10.1016/j.bbalip.2006.08.011.
- Koji Bandoh; Junken Aoki; Akitsu Taira; Masafumi Tsujimoto; Hiroyuki Arai; Keizo Inoue; Lysophosphatidic acid (LPA) receptors of the EDG family are differentially activated by LPA species. FEBS Letters 2000, 478, 159-165, 10.1016/s0014-5793(00)01827-5.
- Koji Bandoh; Junken Aoki; Hiroyuki Hosono; Susumu Kobayashi; Tetsuyuki Kobayashi; Kimiko Murakami-Murofushi; Masafumi Tsujimoto; Hiroyuki Arai; Keizo Inoue; Molecular Cloning and Characterization of a Novel Human G-protein-coupled Receptor, EDG7, for Lysophosphatidic Acid. Journal of Biological Chemistry 1999, 274, 27776-27785, 10.1074/jbc.274.39.27776.
- Rocío Alcántara-Hernández; Aurelio Hernández-Méndez; Gisselle A. Campos-Martínez; Aldo Meizoso Huesca; J. Adolfo García-Sáinz; Phosphorylation and Internalization of Lysophosphatidic Acid Receptors LPA1, LPA2, and LPA3. PLOS ONE 2015, 10, e0140583, 10.1371/journal.pone.0140583.
- James I. Fells; Ryoko Tsukahara; Jianxiong Liu; Gabor Tigyi; Abby L. Parrill; Structure-based drug design identifies novel LPA3 antagonists. Bioorganic & Medicinal Chemistry 2009, 17, 7457-7464, 10.1016/j.bmc.2009.09.022.
- Shimeng Guo; Jiandong Zhang; Shuyong Zhang; Jing Li; A Single Amino Acid Mutation (R104P) in the E/DRY Motif of GPR40 Impairs Receptor Function. PLOS ONE 2015, 10, e0141303, 10.1371/journal.pone.0141303.
- Kate L. White; Matthew T. Eddy; Zhan-Guo Gao; Gye Won Han; Tiffany Lian; Alexander Deary; Nilkanth Patel; Kenneth Jacobson; Vsevolod Katritch; Raymond C. Stevens; et al. Structural Connection between Activation Microswitch and Allosteric Sodium Site in GPCR Signaling. Structure 2018, 26, 259-269.e5, 10.1016/j.str.2017.12.013.
- Mireia Olivella; Gianluigi Caltabiano; Arnau Cordomí; The role of Cysteine 6.47 in class A GPCRs. BMC Structural Biology 2012, 13, 3-3, 10.1186/1472-6807-13-3.
- You-Me Kim; Jeffrey L. Benovic; Differential Roles of Arrestin-2 Interaction with Clathrin and Adaptor Protein 2 in G Protein-coupled Receptor Trafficking. Journal of Biological Chemistry 2002, 277, 30760-30768, 10.1074/jbc.m204528200.
- May M. Paing; Christopher A. Johnston; David P. Siderovski; Joann Trejo; Clathrin Adaptor AP2 Regulates Thrombin Receptor Constitutive Internalization and Endothelial Cell Resensitization. Molecular and Cellular Biology 2006, 26, 3231-42, 10.1128/mcb.26.8.3231-3242.2006.
- Breann L. Wolfe; Joann Trejo; Clathrin-Dependent Mechanisms of G Protein-coupled Receptor Endocytosis. Traffic 2007, 8, 462-470, 10.1111/j.1600-0854.2007.00551.x.
- Owen N. Vickery; Catarina A. Carvalheda; Saheem Zaidi; Andrei Pisliakov; Vsevolod Katritch; Ulrich Zachariae; Intracellular Transfer of Na+ in an Active-State G-Protein-Coupled Receptor. Structure 2017, 26, 171-180.e2, 10.1016/j.str.2017.11.013.
- G. Enrico Rovati; Valérie Capra; Vincent S. Shaw; Rabia U. Malik; Sivaraj Sivaramakrishnan; Richard R. Neubig; The DRY motif and the four corners of the cubic ternary complex model. Cellular Signalling 2017, 35, 16-23, 10.1016/j.cellsig.2017.03.020.
- Qingtong Zhou; Dehua Yang; Meng Wu; Yu Guo; Wangjing Guo; Li Zhong; Xiaoqing Cai; Antao Dai; Wonjo Jang; Eugene I Shakhnovich; et al. Common activation mechanism of class A GPCRs. eLife 2019, 8, e50279, 10.7554/elife.50279.
- Shuguang Yuan; Activation Of G-Protein-Coupled Receptors Correlates With The Formation Of A Continuous Internal Water Pathway. null 2015, 5, 4733, 10.5281/zenodo.32732.
- Xuejun C. Zhang; Ye Zhou; Can Cao; Proton transfer during class-A GPCR activation: do the CWxP motif and the membrane potential act in concert?. Biophysics Reports 2018, 4, 115-122, 10.1007/s41048-018-0056-0.
- Irina S. Moreira; Structural features of the G-protein/GPCR interactions. Biochimica et Biophysica Acta (BBA) - General Subjects 2013, 1840, 16-33, 10.1016/j.bbagen.2013.08.027.
- Daniel Wacker; Raymond Stevens; Bryan L. Roth; How Ligands Illuminate GPCR Molecular Pharmacology. Cell 2017, 170, 414-427, 10.1016/j.cell.2017.07.009.
- Ieva Sutkeviciute; Jean-Pierre Vilardaga; Structural insights into emergent signaling modes of G protein–coupled receptors. Journal of Biological Chemistry 2020, 295, 11626-11642, 10.1074/jbc.rev120.009348.
- Iban Ubarretxena-Belandia; Donald M Engelman; Helical membrane proteins: diversity of functions in the context of simple architecture. Current Opinion in Structural Biology 2001, 11, 370-376, 10.1016/s0959-440x(00)00217-7.
- A E Alewijnse; H Timmerman; E H Jacobs; M J Smit; E Roovers; S Cotecchia; R Leurs; The effect of mutations in the DRY motif on the constitutive activity and structural instability of the histamine H(2) receptor.. Molecular Pharmacology 2000, 57, 890–898, .
- Rong He; Darren D. Browning; Richard D. Ye; Differential roles of the NPXXY motif in formyl peptide receptor signaling.. The Journal of Immunology 2001, 166, 4099-4105, 10.4049/jimmunol.166.6.4099.
- Duane A Chung; Susan M Wade; Carol B Fowler; Danielle D Woods; Paolo B Abada; Henry I Mosberg; Richard R Neubig; Mutagenesis and peptide analysis of the DRY motif in the α2A adrenergic receptor: evidence for alternate mechanisms in G protein-coupled receptors. Biochemical and Biophysical Research Communications 2002, 293, 1233-1241, 10.1016/s0006-291x(02)00357-1.
- X. Edward Zhou; Yuanzheng He; Parker de Waal; Xiang Gao; Yanyong Kang; Ned Van Eps; Yanting Yin; Kuntal Pal; Devrishi Goswami; Thomas A. White; et al. Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors. Cell 2017, 170, 457-469.e13, 10.1016/j.cell.2017.07.002.
- Daniel Mayer; Fred F. Damberger; Mamidi Samarasimhareddy; Miki Feldmueller; Ziva Vuckovic; Tilman Flock; Brian Bauer; Eshita Mutt; Franziska Zosel; Frédéric H. T. Allain; et al. Distinct G protein-coupled receptor phosphorylation motifs modulate arrestin affinity and activation and global conformation. Nature Communications 2019, 10, 1261, 10.1038/s41467-019-09204-y.
- Yuko Fujiwara; Vineet Sardar; Akira Tokumura; Daniel Baker; Kimiko Murakami-Murofushi; Abby Parrill; Gabor Tigyi; Identification of Residues Responsible for Ligand Recognition and Regioisomeric Selectivity of Lysophosphatidic Acid Receptors Expressed in Mammalian Cells. Journal of Biological Chemistry 2005, 280, 35038-35050, 10.1074/jbc.m504351200.
- William J. Valentine; James I. Fells; Donna H. Perygin; Sana Mujahid; Kazuaki Yokoyama; Yuko Fujiwara; Ryoko Tsukahara; James R. Van Brocklyn; Abby L. Parrill; Gabor Tigyi; et al. Subtype-specific Residues Involved in Ligand Activation of the Endothelial Differentiation Gene Family Lysophosphatidic Acid Receptors. Journal of Biological Chemistry 2008, 283, 12175-12187, 10.1074/jbc.m708847200.
- Robert G. Kaye; José W. Saldanha; Zhi-Liang Lu; Edward C. Hulme; Helix 8 of the M1 Muscarinic Acetylcholine Receptor: Scanning Mutagenesis Delineates a G Protein Recognition Site. Molecular Pharmacology 2011, 79, 701-709, 10.1124/mol.110.070177.
- Noel M. Delos Santos; Lidia A. Gardner; Stephen W. White; Suleiman W. Bahouth; Characterization of the Residues in Helix 8 of the Human β1-Adrenergic Receptor That Are Involved in Coupling the Receptor to G Proteins. Journal of Biological Chemistry 2006, 281, 12896-12907, 10.1074/jbc.m508500200.
- John Huynh; Walter Glen Thomas; Marie-Isabel Aguilar; Leonard Keith Pattenden; Role of helix 8 in G protein-coupled receptors based on structure–function studies on the type 1 angiotensin receptor. Molecular and Cellular Endocrinology 2009, 302, 118-127, 10.1016/j.mce.2009.01.002.
- Patricia M. Dijkman; Juan C. Muñoz-García; Steven R. Lavington; Patricia Suemy Kumagai; Rosana Inacio dos Reis; Daniel Yin; Phillip J. Stansfeld; Antonio José Costa-Filho; Anthony Watts; Conformational dynamics of a G protein–coupled receptor helix 8 in lipid membranes. Science Advances 2020, 6, eaav8207, 10.1126/sciadv.aav8207.
- Catalina Ribas; Petronila Penela; Cristina Murga; Alicia Salcedo; Carlota García-Hoz; María Jurado-Pueyo; Ivette Aymerich; Federico Mayor; The G protein-coupled receptor kinase (GRK) interactome: Role of GRKs in GPCR regulation and signaling. Biochimica et Biophysica Acta (BBA) - Biomembranes 2007, 1768, 913-922, 10.1016/j.bbamem.2006.09.019.
- Vsevolod V. Gurevich; Xiufeng Song; Sergey A. Vishnivetskiy; Eugenia V. Gurevich; Enhanced Phosphorylation-Independent Arrestins and Gene Therapy. Organotypic Models in Drug Development 2013, 219, 133-152, 10.1007/978-3-642-41199-1_7.
- Alexey N. Pronin; Christopher V. Carman; Jeffrey L. Benovic; Structure-Function Analysis of G Protein-coupled Receptor Kinase-5. Journal of Biological Chemistry 1998, 273, 31510-31518, 10.1074/jbc.273.47.31510.
- Kenji Watari; Michio Nakaya; Hitoshi Kurose; Multiple functions of G protein-coupled receptor kinases. Journal of Molecular Signaling 2014, 9, 1-1, 10.1186/1750-2187-9-1.
- Matthew T. Drake; Sudha K. Shenoy; Robert J. Lefkowitz; Trafficking of G Protein–Coupled Receptors. Circulation Research 2006, 99, 570-582, 10.1161/01.res.0000242563.47507.ce.
- Eric Reiter; Robert J. Lefkowitz; GRKs and β-arrestins: roles in receptor silencing, trafficking and signaling. Trends in Endocrinology & Metabolism 2006, 17, 159-165, 10.1016/j.tem.2006.03.008.
- Mithu Baidya; Punita Kumari; Hemlata Dwivedi‐Agnihotri; Shubhi Pandey; Madhu Chaturvedi; Tomasz Maciej Stepniewski; Kouki Kawakami; Yubo Cao; Stéphane A Laporte; Jana Selent; et al. Key phosphorylation sites in GPCR s orchestrate the contribution of β‐Arrestin 1 in ERK 1/2 activation. EMBO reports 2020, 21, e49886, 10.15252/embr.201949886.
- Ozge Sensoy; Irina Sousa Moreira; Giulia Morra; Understanding the Differential Selectivity of Arrestins toward the Phosphorylation State of the Receptor. ACS Chemical Neuroscience 2016, 7, 1212-1224, 10.1021/acschemneuro.6b00073.
- Roxanne A. Ally; Kirk L. Ives; Elie Traube; Iman Eltounsi; Pei-Wen Chen; Patrick J. Cahill; James F. Battey; Mark R. Hellmich; Glenn S. Kroog; Agonist- and Protein Kinase C-Induced Phosphorylation Have Similar Functional Consequences for Gastrin-Releasing Peptide Receptor Signaling via Gq. Molecular Pharmacology 2003, 64, 890-904, 10.1124/mol.64.4.890.
- JiHee Kim; SeungKirl Ahn; Xiu-Rong Ren; Erin J. Whalen; Eric Reiter; Huijun Wei; Robert J. Lefkowitz; Functional antagonism of different G protein-coupled receptor kinases for -arrestin-mediated angiotensin II receptor signaling. Proceedings of the National Academy of Sciences 2005, 102, 1442-1447, 10.1073/pnas.0409532102.
- Tilman Flock; Charles N. J. Ravarani; Dawei Sun; A. J. Venkatakrishnan; Melis Kayikci; Christopher G. Tate; Dmitry Veprintsev; M. Madan Babu; Universal allosteric mechanism for Gα activation by GPCRs. Nature 2015, 524, 173-179, 10.1038/nature14663.
- Adrian J. Butcher; Rudi Prihandoko; Kok Choi Kong; Phillip McWilliams; Jennifer M. Edwards; Andrew Bottrill; Sharad Mistry; Andrew B. Tobin; Differential G-protein-coupled Receptor Phosphorylation Provides Evidence for a Signaling Bar Code. Journal of Biological Chemistry 2011, 286, 11506-11518, 10.1074/jbc.m110.154526.
- Aurelio Hernández-Méndez; Rocío Alcántara-Hernández; J. Adolfo García-Sáinz; Lysophosphatidic acid LPA1-3 receptors: signaling, regulation and in silico analysis of their putative phosphorylation sites. Receptors & Clinical Investigation 2014, 1, 10, 10.14800/rci.193.
- Shan Yu; Litao Sun; Yufei Jiao; Leo Tsz On Lee; The Role of G Protein-coupled Receptor Kinases in Cancer. International Journal of Biological Sciences 2017, 14, 189-203, 10.7150/ijbs.22896.
- Joseph B. Black; Richard T. Premont; Yehia Daaka; Feedback regulation of G protein-coupled receptor signaling by GRKs and arrestins. Seminars in Cell & Developmental Biology 2016, 50, 95-104, 10.1016/j.semcdb.2015.12.015.
- David T. Lodowski; Julie A. Pitcher; W. Darrell Capel; Robert J. Lefkowitz; John J. G. Tesmer; Keeping G Proteins at Bay: A Complex Between G Protein-Coupled Receptor Kinase 2 and Gbetagamma. Science 2003, 300, 1256-1262, 10.1126/science.1082348.
- Elisabeth Cassier; Nathalie Gallay; Thomas Bourquard; Sylvie Claeysen; Joël Bockaert; Pascale Crépieux; Anne Poupon; Eric Reiter; Philippe Marin; Franck Vandermoere; et al. Phosphorylation of β-arrestin2 at Thr383 by MEK underlies β-arrestin-dependent activation of Erk1/2 by GPCRs. eLife 2017, 6, e23777, 10.7554/elife.23777.
- Huijun Wei; SeungKirl Ahn; Sudha K. Shenoy; Sadashiva S. Karnik; László Hunyady; Louis M. Luttrell; Robert J. Lefkowitz; Independent -arrestin 2 and G protein-mediated pathways for angiotensin II activation of extracellular signal-regulated kinases 1 and 2. Proceedings of the National Academy of Sciences 2003, 100, 10782-10787, 10.1073/pnas.1834556100.
- Cristina Murga; Ana Ruiz-Gómez; Irene García-Higuera; Chong M. Kim; Jeffrey L. Benovic; Federico Mayor; High Affinity Binding of β-Adrenergic Receptor Kinase to Microsomal Membranes: MODULATION OF THE ACTIVITY OF BOUND KINASE BY HETEROTRIMERIC G PROTEIN ACTIVATION. Journal of Biological Chemistry 1996, 271, 985-994, 10.1074/jbc.271.2.985.
- Takako Shiina; Ken Arai; Shihori Tanabe; Norihiro Yoshida; Tatsuya Haga; Taku Nagao; Hitoshi Kurose; Clathrin Box in G Protein-coupled Receptor Kinase 2. Journal of Biological Chemistry 2001, 276, 33019-33026, 10.1074/jbc.m100140200.
- Sandra Peregrin; Maria Jurado-Pueyo; Pedro M. Campos; Victoria Sanz-Moreno; Ana Ruiz-Gomez; Piero Crespo; Federico Mayor; Cristina Murga; Phosphorylation of p38 by GRK2 at the Docking Groove Unveils a Novel Mechanism for Inactivating p38MAPK. Current Biology 2006, 16, 2042-2047, 10.1016/j.cub.2006.08.083.
- Christopher V. Carman; Larry S. Barak; Chongguang Chen; Lee-Yuan Liu-Chen; James J. Onorato; Scott P. Kennedy; Marc G. Caron; Jeffrey L. Benovic; Mutational Analysis of Gβγ and Phospholipid Interaction with G Protein-coupled Receptor Kinase 2. Journal of Biological Chemistry 2000, 275, 10443-10452, 10.1074/jbc.275.14.10443.
- Suleiman W. Bahouth; Mohammed M. Nooh; Barcoding of GPCR trafficking and signaling through the various trafficking roadmaps by compartmentalized signaling networks. Cellular Signalling 2017, 36, 42-55, 10.1016/j.cellsig.2017.04.015.
- Zhao Yang; Fan Yang; Daolai Zhang; Zhixin Liu; Amy Lin; Chuan Liu; Peng Xiao; Xiao Yu; Jin-Peng Sun; Phosphorylation of G Protein-Coupled Receptors: From the Barcode Hypothesis to the Flute Model. Molecular Pharmacology 2017, 92, 201-210, 10.1124/mol.116.107839.
