Alert! Revision description should not be blank.
Where is the place for revision description?
- yeast
- antimicrobial antagonism
- bioprotection
- wine
- pulcherrimin
- rDNA reticulation
- iron immobilisation
1. Definition
Metschnikowia Kamienski (1899) is a large ascomycetous genus currently comprising 79 species (Mycobank, 04. 2020) but the number or species is continuously growing. The M. pulcherrima clade of the genus contains seven validly described species that share the ability to produce pulcherrimin, a maroon-red pigment (reviewed in [1,2]). These species and the strains closely related to them have broad biotechnological potential for application in various industrial processes. In wine fermentation, these yeasts can modulate the population dynamics of the fermenting yeast communities and produce enzymes and a broad range of compounds that improve the aromatic complexity of the wine (for a review, see [3]).
2. Introduction
3. The M. pulcherrima Clade
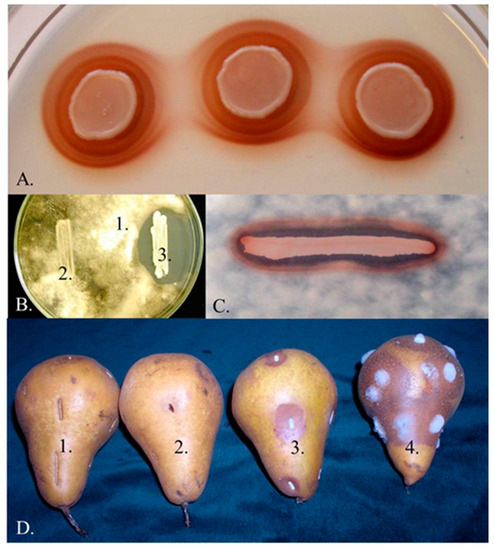
Pulcherrimin-producing Metschnikowia strains are common components of the yeast communities that colonise ripening fruits, flowers (nectar), tree sap fluxes and also frequently occur in fruit juices and fermenting wine (e.g., [2,3,6,9,12,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32]). New yeast isolates producing the characteristic maroon-red pulcherrimin halos around their colonies (Figure 1A) are frequently declared to belong to M. pulcherrima without taking into account that M. pulcherrima is not the only pigment-producing Metschnikowia species. Over the past two decades, five additional species (M. andauensis, M. rubicola, M. shanxiensis, M. sinensis, M. zizyphicola) were validly described and M. fructicola, originally described as a pigment-less species has also turned out to produce pulcherrimin (for a review, see [1]). The intensity of pigment production is variable and highly dependent on the culturing conditions [6,33,34] and probably also on ploidy [35]. The phylogenetic analysis of the barcode sequences of the type strains of the genus clustered these species in a group designated M. pulcherrima clade [1,2].
Recently, two additional species, M. persimmonesis and M. citriensis were proposed to accommodate pulcherrimin-producing strains. The taxonomic name M. persimmonesis was proposed for a single Korean isolate but without providing a complete taxonomic description [27]. The phylogenetic position of the strain is uncertain because its different rDNA barcode sequences (D1/D2, ITS and 18S) show the highest similarities to sequences of the type strains of different species. M. citriensis is based on two strains isolated from citrus leaves [30]. The taxonomic position of these strains is also somewhat obscure, because the authors found them closely related to M. koreensis based on the neighbour-joining analysis of the D1/D2 domains of the 26S rRNA genes, but the M. koreenesis sequence used in the analysis was a direct GenBank submission amplified from a strain for which no taxonomic description is available. The other closest relatives were strains of three pigment-producing members of the M. pulcherrima clade and the non-pigmented M. chrysoperlae, but only one sequence used in the phylogenetic analysis represented a type strain. As previous analyses found M. koreensis separated by a large phylogenetic distance from the M. pulcherrima clade [1,33], the proposed simultaneous close relationship to M. koreensis and the M. pulcherrima clade needs to be revised or reinforced by the analysis or more sequences. Interestingly, when the ITS sequences are examined, the M. citriensis type strain differ more from the other M. citriensis strain than from the M. persimmonesis type strain. Besides, both the D1/D2 and the ITS sequences were cloned, and a phylogenetic analysis based on cloned sequences can easily be misleading in this group of yeast species because of the very high intragenomic diversity of the rDNA repeats (see below). The formation of spheroidal ascospores is another problematic feature of these isolates. M. pulcherrima and its relatives have needle-shape spores [33]. Because of these uncertainties, further examination is required to validate the status of M. persimmonesis and M. citriensis as distinct species. Nevertheless, most properties of their strains and the results of the sequence analyses indicate taxonomic affinity with the M. pulcherrima clade. Many pulcherrimin-producing isolates were not identified at the species level or could not be assigned to any species and were therefore only classified as Metschnikowia sp., M. aff. pulcherrima or M. aff. fructicola. On the other hand, many strains have been classified into these species without presenting sufficient taxonomic evidence. Since pigmentation is an irrelevant property in most biotechnological processes, the strains isolated for industrial purposes are normally not tested for pulcherrimin production. Therefore and because of the sensitivity of pulcherrimin synthesis to the culturing conditions, it is unknown whether pigmentation is a general ability of all strains of the clade.
References
- Lachance, M.A. Metschnikowia: Half tetrads, a regicide and the fountain of youth. Yeast 2016, 33, 563–574. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J.; Basehoar, E.; Ward, T.J. Four new species of Metschnikowia and the transfer of seven Candida species to Metschnikowia and Clavispora as new combinations. Antonie Van Leeuwenhoek 2018, 111, 2017–2035. [Google Scholar] [CrossRef]
- Morata, A.; Loira, I.; Escott, C.; del Fresno, J.M.; Bañuelos, M.A.; Suárez-Lepe, J.A. Applications of Metschnikowia pulcherrima in wine biotechnology. Fermentation 2019, 5, 63. [Google Scholar] [CrossRef]
- Abeln, F.; Chuck, C.J. Achieving a high-density oleaginous yeast culture: Comparison of four processing strategies using Metschnikowia pulcherrima. Biotechnol. Bioeng. 2019, 116, 3200–3214. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Droby, S. Metschnikowia fructicola, a new ascosporic yeast with potential for biocontrol of postharvest fruit rots. Syst. Appl. Microbiol. 2001, 24, 395–399. [Google Scholar] [CrossRef]
- Sipiczki, M. Metschnikowia strains isolated from botrytized grapes antagonize fungal and bacterial growth by iron depletion. Appl. Environ. Microbiol. 2006, 72, 6716–6724. [Google Scholar] [CrossRef]
- Saravanakumar, D.; Ciavorella, A.; Spadaro, D.; Garibaldi, A.; Gullino, M.L. Metschnikowia pulcherrima strain MACH1 outcompetes Botrytis cinerea, Alternaria alternata and Penicillium expansum in apples through iron depletion. Postharvest Biol. Technol. 2008, 49, 121–128. [Google Scholar] [CrossRef]
- Türkel, S.; Korukluoglu, M.; Yavuz, M. Biocontrol activity of the local strain of Metschnikowia pulcherrima on different postharvest pathogens. Biotechnol. Res. Int. 2014, 2014, 397167. [Google Scholar] [CrossRef] [PubMed]
- Kántor, A.; Hutková, J.; Petrová, J.; Hleba, L.; Kačániová, M. Antimicrobial activity of pulcherrimin pigment produced by Metschnikowia pulcherrima against various yeast species. J. Microbiol. Biotechnol. Food Sci. 2015, 5, 282–285. [Google Scholar] [CrossRef]
- Parafati, L.; Vitale, A.; Restuccia, C.; Cirvilleri, G. Biocontrol ability and action mechanism of food-isolated yeast strains against Botrytis cinerea causing post-harvest bunch rot of table grape. Food Microbiol. 2015, 47, 85–92. [Google Scholar] [CrossRef]
- Liu, Y.; Yi, L.; Ruan, C.; Yao, S.; Deng, L.; Zeng, K. Proline increases pigment production to improve oxidative stress tolerance and biocontrol ability of Metschnikowia citriensis. Front. Microbiol. 2019, 10, 1273. [Google Scholar] [CrossRef] [PubMed]
- Pawlikowska, E.; James, S.A.; Breierova, E.; Antolak, H.; Kregiel, D. Biocontrol capability of local Metschnikowia sp. isolates. Antonie Van Leeuwenhoek 2019, 112, 1425–1445. [Google Scholar] [CrossRef] [PubMed]
- Sipiczki, M.; Pfliegler, W.P.; Holb, I.J. Metschnikowia species share a pool of diverse rRNA genes differing in regions that determine hairpin-loop structures and evolve by reticulation. PLoS ONE 2013, 8, e67384. [Google Scholar] [CrossRef]
- Sipiczki, M.; Horvath, E.; Pfliegler, W.P. Birth-and-death evolution and reticulation of ITS segments of Metschnikowia andauensis and Metschnikowia fructicola rDNA repeats. Front. Microbiol. 2018, 9, 1193. [Google Scholar] [CrossRef]
- Piombo, E.; Sela, N.; Wisniewski, M.; Hoffmann, M.; Gullino, M.L.; Allard, M.W.; Levin, E.; Spadaro, D.; Droby, S. Genome sequence, assembly and characterization of two Metschnikowia fructicola strains used as biocontrol agents of postharvest diseases. Front. Microbiol. 2018, 9, 593. [Google Scholar] [CrossRef]
- Venkatesh, A.; Murray, A.L.; Boyle, A.B.; Quinn Farrington, L.; Maher, T.J.; Ó’Gaora, P.; Wolfe, K.H.; O’Brien, C.E.; Butler, G. Draft genome sequence of a highly heterozygous yeast strain from the Metschnikowia pulcherrima subclade, UCD127. Genome Announc. 2018, 6, e00550-18. [Google Scholar] [CrossRef]
- Gore-Lloyd, D.; Sumann, I.; Brachmann, A.O.; Schneeberger, K.; Ortiz-Merino, R.A.; Moreno-Beltrán, M.; Schläfli, M.; Kirner, P.; Santos Kron, A.; Rueda-Mejia, M.P.; et al. Snf2 controls pulcherriminic acid biosynthesis and antifungal activity of the biocontrol yeast Metschnikowia pulcherrima. Mol. Microbiol. 2019, 112, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Molnar, O.; Prillinger, H. Analysis of yeast isolates related to Metschnikowia pulcherrima using the partial sequences of the large subunit rDNA and the actin gene; description of Metschnikowia andauensis sp. nov. Syst. Appl. Microbiol. 2005, 28, 717–726. [Google Scholar] [CrossRef]
- Xue, M.L.; Zhang, L.Q.; Wang, Q.M.; Zhang, J.S.; Bai, F.Y. Metschnikowia sinensis sp. nov., Metschnikowia zizyphicola sp. nov. and Metschnikowia shanxiensis sp. nov., novel yeast species from jujube fruit. Int. J. Syst. Evol. Microbiol. 2006, 56, 2245–2250. [Google Scholar] [CrossRef]
- Sipiczki, M. Overwintering of vineyard yeasts: Survival of interacting yeast communities in grapes mummified on vines. Front. Microbiol. 2016, 7, 212. [Google Scholar] [CrossRef]
- Türkel, S.; Ener, B. Isolation and characterization of new Metschnikowia pulcherrima strains as producers of the antimicrobial pigment pulcherrimin. Z. Naturforsch. C J. Biosci. 2009, 64, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Spadaro, D.; Garibaldi, A.; Gullino, M.L. Selection and evaluation of new antagonists for their efficacy against postharvest brown rot of peaches. Postharvest Biol. Technol. 2009, 55, 174–181. [Google Scholar] [CrossRef]
- Vadkertiova, R.; Molnarova, J.; Vranova, D.; Slavikova, E. Yeasts and yeast-like organisms associated with fruits and blossoms of different fruit trees. Can. J. Microbiol. 2012, 58, 1344–1352. [Google Scholar] [CrossRef] [PubMed]
- Csutak, O.; Vassu, T.; Cornea, P.; Grebenisan, I. Genetic characterization of two new Metschnikowia strains with antifungal activity. Rom. Biotech. Lett. 2007, 12, 3175–3182. [Google Scholar]
- Csutak, O.; Vassu, T.; Sarbu, I.; Stoica, I.; Cornea, P. Antagonistic activity of three newly isolated yeast strains from the surface of fruits. Food Technol. Biotechnol. 2013, 51, 70–77. [Google Scholar]
- Hadwiger, L.A.; McDonel, H.; Glawe, D. Wild yeast strains as prospective candidates to induce resistance against potato late blight (Phytophthora infestans). Am. J. Potato Res. 2015, 92, 379–386. [Google Scholar] [CrossRef]
- Kang, Y.M.; Choi, J.E.; Komakech, R.; Park, J.H.; Kim, D.W.; Cho, K.M.; Kang, S.M.; Choi, S.H.; Song, K.C.; Ryu, C.M.; et al. Characterization of a novel yeast species Metschnikowia persimmonesis KCTC 12991BP (KIOM G15050 type strain) isolated from a medicinal plant, Korean persimmon calyx (Diospyros kaki Thumb). AMB Express 2017, 7, 199. [Google Scholar] [CrossRef]
- Pawlikowska, E.; Kręgiel, D. Enzymatic profiles and antimicrobial activity of the yeast Metschnikowia pulcherrima. Acta Innov. 2017, 23, 17–24. [Google Scholar]
- Liu, Y.; Wang, W.; Zhou, Y.; Yaoa, S.; Denga, L.; Zeng, K. Isolation, identification and in vitro screening of Chongqing orangery yeasts for the biocontrol of Penicillium digitatum on citrus fruit. Biol. Control 2017, 110, 18–24. [Google Scholar] [CrossRef]
- Liu, Y.; Yao, S.; Deng, L.; Ming, J.; Zeng, K. Metschnikowia citriensis sp. nov., a novel yeast species isolated from leaves with potential for biocontrol of postharvest fruit rot. Biol. Control 2018, 125, 15–19. [Google Scholar] [CrossRef]
- Tian, Y.Q.; Li, W.; Jiang, Z.T.; Jing, M.M.; Shao, Y.Z. The preservation effect of Metschnikowia pulcherrima yeast on anthracnose of postharvest mango fruits and the possible mechanism. Food Sci. Biotechnol. 2017, 27, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, C.; Lage, P.; Esteves, M.; Chambel, L.; Mendes-Faia, A.; Mendes-Ferreira, A. Molecular and phenotypic characterization of Metschnikowia pulcherrima strains from Douro wine region. Fermentation 2018, 4, 8. [Google Scholar] [CrossRef]
- Lachance, M.A. Metschnikowia Kamienski (1899). In The Yeasts. A Taxonomic Study; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 575–620. [Google Scholar]
References
- Lachance, M.A. Metschnikowia: Half tetrads, a regicide and the fountain of youth. Yeast 2016, 33, 563–574. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J.; Basehoar, E.; Ward, T.J. Four new species of Metschnikowia and the transfer of seven Candida species to Metschnikowia and Clavispora as new combinations. Antonie Van Leeuwenhoek 2018, 111, 2017–2035. [Google Scholar] [CrossRef]
- Morata, A.; Loira, I.; Escott, C.; del Fresno, J.M.; Bañuelos, M.A.; Suárez-Lepe, J.A. Applications of Metschnikowia pulcherrima in wine biotechnology. Fermentation 2019, 5, 63. [Google Scholar] [CrossRef]
- Abeln, F.; Chuck, C.J. Achieving a high-density oleaginous yeast culture: Comparison of four processing strategies using Metschnikowia pulcherrima. Biotechnol. Bioeng. 2019, 116, 3200–3214. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Droby, S. Metschnikowia fructicola, a new ascosporic yeast with potential for biocontrol of postharvest fruit rots. Syst. Appl. Microbiol. 2001, 24, 395–399. [Google Scholar] [CrossRef]
- Sipiczki, M. Metschnikowia strains isolated from botrytized grapes antagonize fungal and bacterial growth by iron depletion. Appl. Environ. Microbiol. 2006, 72, 6716–6724. [Google Scholar] [CrossRef]
- Saravanakumar, D.; Ciavorella, A.; Spadaro, D.; Garibaldi, A.; Gullino, M.L. Metschnikowia pulcherrima strain MACH1 outcompetes Botrytis cinerea, Alternaria alternata and Penicillium expansum in apples through iron depletion. Postharvest Biol. Technol. 2008, 49, 121–128. [Google Scholar] [CrossRef]
- Türkel, S.; Korukluoglu, M.; Yavuz, M. Biocontrol activity of the local strain of Metschnikowia pulcherrima on different postharvest pathogens. Biotechnol. Res. Int. 2014, 2014, 397167. [Google Scholar] [CrossRef] [PubMed]
- Kántor, A.; Hutková, J.; Petrová, J.; Hleba, L.; Kačániová, M. Antimicrobial activity of pulcherrimin pigment produced by Metschnikowia pulcherrima against various yeast species. J. Microbiol. Biotechnol. Food Sci. 2015, 5, 282–285. [Google Scholar] [CrossRef]
- Parafati, L.; Vitale, A.; Restuccia, C.; Cirvilleri, G. Biocontrol ability and action mechanism of food-isolated yeast strains against Botrytis cinerea causing post-harvest bunch rot of table grape. Food Microbiol. 2015, 47, 85–92. [Google Scholar] [CrossRef]
- Liu, Y.; Yi, L.; Ruan, C.; Yao, S.; Deng, L.; Zeng, K. Proline increases pigment production to improve oxidative stress tolerance and biocontrol ability of Metschnikowia citriensis. Front. Microbiol. 2019, 10, 1273. [Google Scholar] [CrossRef] [PubMed]
- Pawlikowska, E.; James, S.A.; Breierova, E.; Antolak, H.; Kregiel, D. Biocontrol capability of local Metschnikowia sp. isolates. Antonie Van Leeuwenhoek 2019, 112, 1425–1445. [Google Scholar] [CrossRef] [PubMed]
- Sipiczki, M.; Pfliegler, W.P.; Holb, I.J. Metschnikowia species share a pool of diverse rRNA genes differing in regions that determine hairpin-loop structures and evolve by reticulation. PLoS ONE 2013, 8, e67384. [Google Scholar] [CrossRef]
- Sipiczki, M.; Horvath, E.; Pfliegler, W.P. Birth-and-death evolution and reticulation of ITS segments of Metschnikowia andauensis and Metschnikowia fructicola rDNA repeats. Front. Microbiol. 2018, 9, 1193. [Google Scholar] [CrossRef]
- Piombo, E.; Sela, N.; Wisniewski, M.; Hoffmann, M.; Gullino, M.L.; Allard, M.W.; Levin, E.; Spadaro, D.; Droby, S. Genome sequence, assembly and characterization of two Metschnikowia fructicola strains used as biocontrol agents of postharvest diseases. Front. Microbiol. 2018, 9, 593. [Google Scholar] [CrossRef]
- Venkatesh, A.; Murray, A.L.; Boyle, A.B.; Quinn Farrington, L.; Maher, T.J.; Ó’Gaora, P.; Wolfe, K.H.; O’Brien, C.E.; Butler, G. Draft genome sequence of a highly heterozygous yeast strain from the Metschnikowia pulcherrima subclade, UCD127. Genome Announc. 2018, 6, e00550-18. [Google Scholar] [CrossRef]
- Gore-Lloyd, D.; Sumann, I.; Brachmann, A.O.; Schneeberger, K.; Ortiz-Merino, R.A.; Moreno-Beltrán, M.; Schläfli, M.; Kirner, P.; Santos Kron, A.; Rueda-Mejia, M.P.; et al. Snf2 controls pulcherriminic acid biosynthesis and antifungal activity of the biocontrol yeast Metschnikowia pulcherrima. Mol. Microbiol. 2019, 112, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Molnar, O.; Prillinger, H. Analysis of yeast isolates related to Metschnikowia pulcherrima using the partial sequences of the large subunit rDNA and the actin gene; description of Metschnikowia andauensis sp. nov. Syst. Appl. Microbiol. 2005, 28, 717–726. [Google Scholar] [CrossRef]
- Xue, M.L.; Zhang, L.Q.; Wang, Q.M.; Zhang, J.S.; Bai, F.Y. Metschnikowia sinensis sp. nov., Metschnikowia zizyphicola sp. nov. and Metschnikowia shanxiensis sp. nov., novel yeast species from jujube fruit. Int. J. Syst. Evol. Microbiol. 2006, 56, 2245–2250. [Google Scholar] [CrossRef]
- Sipiczki, M. Overwintering of vineyard yeasts: Survival of interacting yeast communities in grapes mummified on vines. Front. Microbiol. 2016, 7, 212. [Google Scholar] [CrossRef]
- Türkel, S.; Ener, B. Isolation and characterization of new Metschnikowia pulcherrima strains as producers of the antimicrobial pigment pulcherrimin. Z. Naturforsch. C J. Biosci. 2009, 64, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Spadaro, D.; Garibaldi, A.; Gullino, M.L. Selection and evaluation of new antagonists for their efficacy against postharvest brown rot of peaches. Postharvest Biol. Technol. 2009, 55, 174–181. [Google Scholar] [CrossRef]
- Vadkertiova, R.; Molnarova, J.; Vranova, D.; Slavikova, E. Yeasts and yeast-like organisms associated with fruits and blossoms of different fruit trees. Can. J. Microbiol. 2012, 58, 1344–1352. [Google Scholar] [CrossRef] [PubMed]
- Csutak, O.; Vassu, T.; Cornea, P.; Grebenisan, I. Genetic characterization of two new Metschnikowia strains with antifungal activity. Rom. Biotech. Lett. 2007, 12, 3175–3182. [Google Scholar]
- Csutak, O.; Vassu, T.; Sarbu, I.; Stoica, I.; Cornea, P. Antagonistic activity of three newly isolated yeast strains from the surface of fruits. Food Technol. Biotechnol. 2013, 51, 70–77. [Google Scholar]
- Hadwiger, L.A.; McDonel, H.; Glawe, D. Wild yeast strains as prospective candidates to induce resistance against potato late blight (Phytophthora infestans). Am. J. Potato Res. 2015, 92, 379–386. [Google Scholar] [CrossRef]
- Kang, Y.M.; Choi, J.E.; Komakech, R.; Park, J.H.; Kim, D.W.; Cho, K.M.; Kang, S.M.; Choi, S.H.; Song, K.C.; Ryu, C.M.; et al. Characterization of a novel yeast species Metschnikowia persimmonesis KCTC 12991BP (KIOM G15050 type strain) isolated from a medicinal plant, Korean persimmon calyx (Diospyros kaki Thumb). AMB Express 2017, 7, 199. [Google Scholar] [CrossRef]
- Pawlikowska, E.; Kręgiel, D. Enzymatic profiles and antimicrobial activity of the yeast Metschnikowia pulcherrima. Acta Innov. 2017, 23, 17–24. [Google Scholar]
- Liu, Y.; Wang, W.; Zhou, Y.; Yaoa, S.; Denga, L.; Zeng, K. Isolation, identification and in vitro screening of Chongqing orangery yeasts for the biocontrol of Penicillium digitatum on citrus fruit. Biol. Control 2017, 110, 18–24. [Google Scholar] [CrossRef]
- Liu, Y.; Yao, S.; Deng, L.; Ming, J.; Zeng, K. Metschnikowia citriensis sp. nov., a novel yeast species isolated from leaves with potential for biocontrol of postharvest fruit rot. Biol. Control 2018, 125, 15–19. [Google Scholar] [CrossRef]
- Tian, Y.Q.; Li, W.; Jiang, Z.T.; Jing, M.M.; Shao, Y.Z. The preservation effect of Metschnikowia pulcherrima yeast on anthracnose of postharvest mango fruits and the possible mechanism. Food Sci. Biotechnol. 2017, 27, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, C.; Lage, P.; Esteves, M.; Chambel, L.; Mendes-Faia, A.; Mendes-Ferreira, A. Molecular and phenotypic characterization of Metschnikowia pulcherrima strains from Douro wine region. Fermentation 2018, 4, 8. [Google Scholar] [CrossRef]
- Lachance, M.A. Metschnikowia Kamienski (1899). In The Yeasts. A Taxonomic Study; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 575–620. [Google Scholar]
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms8071029
