Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Virology
Phage lytic proteins are a clinically advanced class of novel enzyme-based antibiotics, so-called enzybiotics.
- phage lytic proteins
- endolysins
- machine learning
- biological database
- conserved protein domains
Bacteriophages are infective viral particles targeting bacterial cells. During their lytic replication cycle, phages face twice the challenge to cross the bacterial cell wall of their host [1]. Therefore, phages make use of two types of phage lytic proteins: virion-associated lysins (VALs) and endolysins [2]. A VAL is part of the virion and forms a local and small pore in the peptidoglycan layers at the site of infection, while the cell remains intact. The phage genome is ejected through this pore. VALs are mostly a structural part of the virion but can also occur as internal capsid proteins [3,4]. Endolysins are responsible for the massive degradation of the peptidoglycan layer at the end of the lytic cycle. In the canonical phage lysis system, they accumulate in large numbers in the cytoplasm and their release into the periplasmic space is timed by pore-forming holins in the cytoplasmic membrane. The sudden degradation of the peptidoglycan, assisted by the high osmotic pressure inside the cell, causes cell lysis and the concomitant release of newly matured phage particles [5,6].
Phage lytic proteins comprise one or more functional domains categorized into two classes: enzymatically active domains (EADs) and cell wall binding domains (CBDs) [7]. Additionally, VALs contain domains with a function other than peptidoglycan degradation such as structural anchoring to the viral particle. At the biochemical level all phage lytic proteins have the same purpose, i.e., peptidoglycan degradation, yet variation in environment and host has led to highly diverse domains and architectures [1,3,4,8]. This variety largely springs from the diversity of peptidoglycan chemotypes among bacterial species, along with the high diversity of secondary cell wall-associated carbohydrate polymers [9,10]. Endolysins have either a globular (one EAD) or modular architecture (multiple domains; at least one EAD and optionally one or more CBDs) [11]. CBDs target the glycan or peptide moieties of peptidoglycan, or specific components of (lipo)teichoic acids, increasing the proximity of EAD to its substrate [12,13]. Five classes of EADs are distinguished based on the bond they cleave in peptidoglycan (Figure 1). (i) N-acetylmuramoyl-L-alanine amidases (EC 3.5.1.28) hydrolyze the amide bond between N-acetylmuramic acid (MurNAc) and L-alanine residues, effectively cleaving the peptide moiety from the glycan strand. (ii) N-acetyl-β-D-glucosaminidases catalyze the hydrolysis of glycosidic β-1,4 linkages between N-acetylglucosamine (GlcNac) and MurNAc. (iii) N-acetyl-β-D-muramidases (EC 3.2.1.17) and (iv) lytic transglycosylases (EC 4.2.2.n2) cleave these linkages between MurNAc and GlcNAc. N-acetyl-β-D-glucosaminidases and N-acetyl-β-D-muramidases are glycosidases (EC 3.2.1.-) that use a hydrolytic mechanism resulting in a terminal reducing GlcNAc or MurNAc residue, respectively. Lytic transglycosylases, on the other hand, use an intramolecular mechanism that creates a 1,6-anhydro bond at the MurNAc residue. Peptidases (EC 3.4.-.-) cleave the bond between two amino acids within the peptidoglycan stem peptide, cross-link or cross-bridge [14]. The specific epitope and chemical bond targeted by the CBD and the EAD, respectively, brings about a well-defined spectrum of activity for phage lytic proteins [15,16]. The modularity of phage lytic proteins, along with the diverse range of domains, has been exploited by protein engineers to modulate the specificity, activity and solubility by domain swapping [11,17].
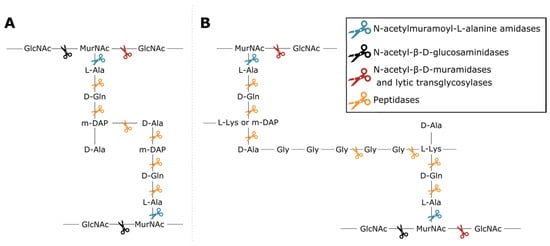
Figure 1. Cleavage sites of the different enzymatic classes of EADs on the primary structures of two common peptidoglycan types. (A) peptidoglycan chemotype A1γ is the most common in Gram-negative bacteria; (B) peptidoglycan chemotype A3, either α or γ depending on the presence of L-Lys or mDAP in the third position of the peptide subunit, is a type example for Gram-positive bacteria such as Staphylococcus aureus [10]. The colored scissors indicate cleavage sites of different classes of EADs. GlcNAc and MurNAc in the glycan strands of each structure refer to N-acetylglucosamine and N-acetylmuramic acid, respectively.
As early as 1957, it was observed that phage lytic proteins can cause “lysis from without” upon exogenous addition to bacteria [18,19]. It was not until 2001 that their use as enzyme-based antibacterial agents, coined “enzybiotics”, was demonstrated in a murine model against Gram-positive bacteria [20]. The presence of an outer membrane was initially prohibitive for the use of phage lytic proteins as enzybiotics against Gram-negative bacteria, but meanwhile various protein engineering approaches have been developed to overcome this barrier, including the use of outer membrane permeabilizing peptides (Artilysins®) [11], bacteriocin domains (lysocins) [21] and phage receptor binding proteins (Innolysins) [22]. Today, enzybiotics are considered the most advanced alternative class of antibacterials under clinical investigation [23]. They offer a necessary response to the alarming threat of antibiotic resistance across global health care systems. Their mode of action is fundamentally different from any existing class of antibiotics. Enzybiotics actively degrade the peptidoglycan component without the need for an active bacterial metabolism, unlike classic antibiotics. This is reflected in a faster cell death and effectiveness against metabolically inactive cells like persisters. In addition, their spectrum is typically narrower (genus, species or strain level) compared to classic antibiotics, causing less harm to beneficial microflora. Finally, the conserved molecular target makes enzybiotics less prone to the inevitable fate of many traditional antibiotics: the emergence and spread of resistance mechanisms [11,24]. A growing community of researchers and companies is therefore investigating their applications, including clinical trials, and engineering their properties to kill a broad diversity of bacteria [8]. Phage lytic proteins have the inherent potential to be developed against any bacterial species, thus representing an unprecedented extensive class of antibiotics with narrow spectrum.
This entry is adapted from the peer-reviewed paper 10.3390/v13071240
This entry is offline, you can click here to edit this entry!
