Host genetic diversity tends to limit disease spread in nature and buffers populations against epidemics. Genetic diversity in wildlife is expected to receive increasing attention in contexts related to disease transmission and human health.
- genetic diversity
- disease spread
- tuberculosis
1. Introduction
2. Impact of Infectious Diseases on Host Populations
Throughout generations, interactions with pathogens produce evolutionary changes in host populations [39][72]. In a host population, pathogen infections favor genotypes with higher resistance (ability to limit pathogen burden [73][74]) or tolerance (ability to limit disease severity induced by a given pathogen burden [73]). In addition to changes in resistance or tolerance, diseases can have other impacts on host populations: population reduction, changes of age structure, alteration on life-history parameters, or effects on genetic diversity [75]. However, the presence of pathogens can also cause changes to host behavior.
For instance, individuals can avoid infected mates to reduce pathogen transmission [76][77][78]. However, host–pathogen interactions have induced evolutionary processes that are responsible for the functioning of other mate choice-related behaviors. On the other hand, individuals (mainly females [79][80]) can choose genetically dissimilar mates to promote genetic diversity of descendants and, hence, their capacity to resist or tolerate pathogens [81][82][83]. However, the existence of infectious diseases might boost individuals to choose mates with the same level of infection, a behavior that tends to favor genetically similar mating and loss of population genetic diversity [84][85].
Studies have also investigated the effects of infectious diseases on dispersal behavior that, in turn, influence the genetic structure of populations [86][87]. Demographic declines that follow disease outbreaks increase resource availability and decrease dispersal advantages. Consequently, the low need for dispersal reduces gene flow and enhances genetic differentiation. However, the expected reduction in genetic diversity as a consequence of low dispersal might be counteracted by the effect of balancing selection acting on immune genes during the disease outbreak [88].
Pathogen– host coevolutionary dynamics may be characterized by fluctuating selection (FS), where host genotypes may be at any moment more resistant to contemporary, compared to past or future pathogens, or by arms races (AR), where both hosts and pathogens tend to increase resistance/infectivity over time [89]. The FS dynamic is based on specialized interactions and, hence, it is typical of spatially structured environments. Mixing locally adapted phenotypes may shift the coevolutionary interactions from FS to AR, due to exposure to a higher range of genotypes selected for a wider range of resistance/infectivity in both coevolutionary counterparts [90].
Since spatial variation causes the evolution of locally adaptive immunity, individuals might tend to reduce the contact or refuse mating with genetically different conspecifics harboring dangerous pathogens [91][92]. Genetically different individuals might tolerate pathogens for which local immune systems might not be prepared. They also include genes related to immune system that have not been selected under the local pathogen–host coevolutionary dynamics. These characteristics might make local individuals reluctant to contact and mate with genetically different individuals proceeding from distant populations.
3. Ungulates as Hosts
Among mammals, ungulates (a paraphyletic group that includes Artiodactyla and Perissodactyla orders) include a high proportion of wild species with zoonotic diseases. [93] found that 32% of wild ungulate species (73/247 species) were zoonotic hosts. The high rates of disease transmission from wild ungulates to human have been mainly driven by our contact with these species throughout human history [94][95][96]. In addition to promoting species conservation, the maintenance of high levels of genetic diversity in wild ungulates should reduce risks regarding the emergence of infectious diseases, these risks being some of the most important threats to human health and the global economy [97][98][99].
Genetic diversity of ungulates is being altered by human-mediated processes acting on gene flow or effective size of populations. Due to hunting, competition with livestock, and lost habitat, many ungulates occur in small or bottleneck populations [100][101]. Sex-biased harvesting changes population structures and reduces effective population sizes [102][103]. These processes tend to decrease ungulate genetic diversity and, hence, affect species conservation and the probability of infectious disease emergence and spread.
Despite human-mediated alterations of gene flows and effective population sizes, studies addressing genetic diversity of ungulate populations focus primarily on conservation prospects, rather than on its effects on disease emergence and spread (Figure 1). This search showed studies that relate host genetic diversity to the spread of infectious diseases [104] (see below); studies on population genetic structure that highlight the importance of genetic diversity on disease emergence, spread, or development [105][106][107][108][109][110][111][112][113][114]; studies that analyze genetic loci or genetic metrics related to the ability of individuals and population to deal with pathogens and diseases [115][116][117]; studies that associate low genetic diversity with the presence of non-infectious diseases, alterations, or distinctive traits [118][119][120][121][122][123]; and studies on heterozygosity–fitness correlations that show expected [124] or unexpected results [125][126]. Therefore, a notable disconnection appears between studies treating ungulate genetic diversity and those dealing with infectious diseases in ungulates. The emergence and spread of infectious diseases, as well as their threats for human health and economy, might be explicitly added to conservation arguments when dealing with genetic diversity of wildlife.
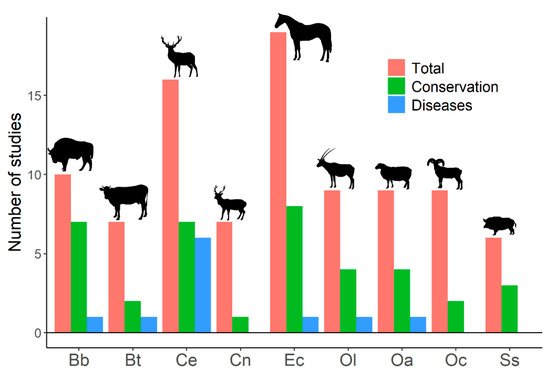
Figure 1. Number of published studies on genetic diversity for the most frequently studied ungulates. Results from a search on the Web of Science with the following search terms: genetic diversity, inbreeding, and ungulates (217 studies were obtained). Studies on genetic diversity of ungulate populations published in scientific journals were selected (204 papers). Total: number of studies on genetic diversity of ungulate populations published in scientific journals. Conservation: number of studies that explicitly related genetic diversity to conservation (papers in which the word ‘conservation’ appeared in the title, abstract, or the name of the journal). Diseases: number of studies that explicitly associated genetic diversity with diseases (papers in which the title, abstract, or name of the journal used at least one of the following terms: ‘disease’, ‘pathogen’, ‘parasite’, any variation of ‘immunity’, or the name of any disease). Bb: Bison bonasus, Bt: Bos taurus, Ce: Cervus elaphus, Cn: Cervus nippon, Ec: Equus caballus, Ol: Oryx leucoryx, Oa: Ovis aries, Oc: Ovis canadensis, Ss: Sus scrofa. The search was last consulted on 15 April 2021.
4. Factors Affecting Wild Boar and Red Deer Genetic Diversity. Recommendations to Confront TB
Wild boar and red deer are important game species in Spain, where most populations are in private hunting estates (typically 750–3000 ha). In these private estates, wild boar and red deer can coexist with other wild ungulates, such as fallow deer, or domestic ungulates, such as cattle [127][128]. These management actions alter ecology and behavior of individuals that, in turn, affect gene flow and effective population size. Consequently, some populations can present low levels of genetic diversity or high inbreeding, reference [104][108][129][130][131][132][133][134][135] which are associated with low antler development in red deer [121] and high predisposition to TB progression in both species [104][129][130].
A management action, presumably affecting genetic diversity, is the placement of high perimetral fences around the estates to maintain wild ungulates inside the owned land [136][137][138]. As a result of this practice, in some of the properties, we can find two types of hunting estates: open (without perimetral fences) and fenced (with perimetral fences). Additionally, small effective population sizes at the moment of fence placement might cause a founder effect with important consequences on genetic diversity and inbreeding. The lack of gene flow and founder effects might make wild boar and red deer in fenced estates present low levels of genetic diversity and might make potentially dangerous populations.
Additionally, studies comparing red deer populations in open and fenced estates have found that there are not differences in genetic diversity between both types of estates [131][134][135]. Fences might allow a certain degree of individual movements among estates and, hence, genetic diversity in fenced estates can be maintained. In addition to fence permeability, behavioral processes might avoid the loss of genetic diversity in both species [139][140][141] (see below). In spite of the existence of these processes tending to maintain reservoir genetic diversity, fenced estates, mainly those in which wild reservoirs and livestock coexist, bear potential risks that should be monitored.
Owners and managers might increase genetic diversity and reduce inbreeding in fenced estates by translocating individuals from different populations [142]. However, translocations imply the existence of additional risks that might overcome their benefits in maintaining reservoir genetic diversity. Moreover, translocated individuals swamp the genetic variation of the native populations and, hence, increase homogenization and reduce genetic diversity of a species scale [143][144][145]. In order to minimize the disadvantages of translocations, owners and managers should select individuals from populations that occur in nearby, similar habitats, and with low genetic divergence [146][147].
In open estates, managers promote the harvesting of the maximum number of males before they are hunted by neighbors. The low rate of migration rates of males among open estates might hinder the maintenance of genetic diversity and explain why open and fenced estates do not present different levels of genetic variation. Therefore, genetic diversity in open estates can be, to some extent, maintained by female dispersal. Nevertheless, a reduction in hunting harvesting over males can be recommended to equilibrate population structure and recover natural dispersal of males.
In addition to fences and population structure, landscape might also influence dispersal and gene flow in wild boar and red deer. For red deer, however, landscape features significantly affect movements [148]. In Spain, forest continuity has been shown to favor red deer movements and dispersal [134]. Therefore, to avoid loss of generic diversity in red deer populations, refuge (forest) continuity might be recommended to facilitate individual movements between open estates.
In addition to processes related to dispersal, game management affects the mating system of Spanish red deer and wild boar populations. However, high levels of mate competition in fenced estates favor the success of those males with higher levels of genetic diversity [141]. It is worth highlighting that the genetic diversity contributed by males tends to be higher than that transmitted by females in populations with high levels of mate competition [141]. The higher effective population size and genetic diversity contributed by males might help to explain why red deer populations in fenced estates have similar genetic diversity than that in open estates.
Another mating system related behavior that can be affected by game management is dissimilar mating. In open estates, the low proportion of males, that are mainly young and philopatric, hinders the action of dissimilar mating. Therefore, dissimilar mating in fenced estates, where the proportion of adult males is high, might favor genetic diversity conservation and might also help to explain the lack of differences in genetic diversity between fenced and open estates. However, this argument loses importance in those open estates where females disperse to avoid inbreeding [138].
Inbreeding depression is an important selective pressure affecting behavioral processes related to mating system [149][150]. These processes tend to compensate the loss of genetic diversity of populations and their action depends on the existence of equilibrated population structures. Male-biased hunting in red deer causes altered population structures that hinder the action of behaviors favoring genetic diversity conservation. Whenever possible, hunting regimes favoring equilibrated population structures in red deer populations might be recommended.
In wild boar, mating system related processes affecting genetic diversity have been found. Firstly, multiple paternity (different males siring offspring within the same litter) tends to maintain genetic diversity [151][152] and it has been found in wild boar populations [139]. Multiple paternity and male mate competition, both tending to favor genetic diversity conservation, are expected to act mainly in equilibrated population structures with high proportion of adult males. Nevertheless, management actions ensuring equilibrated population structures might be recommended to favor genetic diversity conservation.
This entry is adapted from the peer-reviewed paper 10.3390/ani11061630
References
- Kohn, M.H.; Murphy, W.J.; Ostrander, E.A.; Wayne, R. Genomics and conservation genetics. Trends Ecol. Evol. 2006, 21, 629–637.
- Frankel, O.H. Genetic conservation: Our evolutionary responsibility. Genetics 1974, 78, 53–65.
- Frankel, O.H. Variation, the essence of life. Proc. Linn. Soc. N. S. W. 1970, 95, 158–169.
- Charlesworth, B.; Charlesworth, D. The genetic basis of inbreeding depression. Genet. Res. 1999, 74, 329–340.
- Frankel, O.H.; Soulé, M.E. Conservation and Evolution; Cambridge University Press: Cambridge, UK, 1981.
- Neaves, L.E.; Eales, J.; Whitlock, R.; Hollingsworth, P.M.; Buerke, T.; Pullin, A.S. The fitness consequences of inbreeding in natural populations and their implications for species conservation—A systematic map. Environ. Evid. 2015, 4, 5.
- Frankham, R. Genetic adaptation to captivity in species conservation programs. Mol. Ecol. 2008, 17, 325–333.
- Frankham, R. Genetics and extinction. Biol. Conserv. 2005, 126, 131–140.
- Amos, W.; Balmford, A. When does conservation genetics matter? Heredity 2001, 87, 257–265.
- O’Brien, S.J. A role for molecular genetics in biological conservation. Proc. Natl. Scad. Sci. USA 1994, 91, 5748–5755.
- Morse, S.S.; Mazet, J.A.; Woolhouse, M.; Parrish, C.R.; Carroll, D.; Karesh, W.B.; Zambrana-Torrelio, C.; Lipkin, W.I.; Daszak, P. Prediction and prevention of the next pandemic zoonosis. Lancet 2012, 380, 1956–1965.
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993.
- Kruse, H.; Kirkemo, A.-M.; Handeland, K. Wildlife as source of zoonotic Infections. Emerg. Infect. Dis. 2004, 10, 2067–2072.
- Taylor, L.H.; Latham, S.M.; Woolhouse, M.E.J. Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 983–989.
- Allen, T.; Murray, K.A.; Zambrana-Torrelio, C.; Morse, S.S.; Rondinini, C.; Di Marco, M.; Breit, N.; Olival, K.J.; Daszak, P. Global hotspots and correlates of emerging zoonotic diseases. Nat. Commun. 2017, 8, 1124.
- Heymann, D.L.; Chen, L.; Takemi, K.; Fidler, D.P.; Tappero, J.W.; Thomas, M.J.; Kenyon, T.A.; Frieden, T.; Yach, D.; Nishtar, S.; et al. Global health security: The wider lessons from the west African Ebola virus disease epidemic. Lancet 2015, 385, 1884–1901.
- Morens, D.M.; Fauci, A.S. Emerging infectious diseases in 2012: 20 years after the institute of medicine report. Mbio 2012, 3, e00494-12.
- Lu, H.; Stratton, C.W.; Tang, Y. Outbreak of pneumonia of unknown etiology in Wuhan China: The mystery and the miracle. J. Med. Virol. 2020, 93, 401–402.
- McKibbin, W.; Fernando, R. The Global Macroeconomic Impacts of COVID-19: Seven Scenarios. Asian Econ. Pap. 2021, 20, 1–30.
- Sohrabi, C.; Alsafi, Z.; O’Neill, N.; Khan, M.; Kerwan, A.; Al-Jabir, A.; Iosifidis, C.; Agha, R. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int. J. Surg. 2020, 76, 71–76.
- Pike, J.; Bogich, T.L.; Elwood, S.; Finnoff, D.C.; Daszak, P. Economic optimization of a global strategy to reduce the pandemic threat. Proc. Natl. Acad. Sci. USA 2014, 111, 18519–18523.
- Quéméré, E.; Rossi, S.; Petit, E.; Marchard, P.; Merlet, J.; Game, Y.; Galan, M.; Gilot-Fromont, E. Genetic epidemiology of the Alpine ibex reservoir of persistent and virulent brucellosis outbreak. Sci. Rep. 2020, 10, 4400.
- Portanier, E.; Garel, M.; Devillard, S.; Maillard, D.; Poissant, J.; Galan, M.; Benabed, S.; Poirel, M.T.; Duhayer, J.; Itty, C.; et al. Both candidate gene and neutral genetic diversity correlate with parasite resistance in female Mediterranean mouflon. BMC Ecol. 2019, 19, 12.
- Mitchell, J.; Vitikainen, E.I.K.; Wells, D.A.; Cant, M.A.; Nichols, H.J. Heterozygosity but not inbreeding coefficient predicts parasite burdens in the banded mongoose. J. Zool. 2017, 302, 32–39.
- Benavides, M.V.; Sonstegard, T.S.; Van Tassell, C. Genomic regions associated with sheep resistance to gastrointestinal nematodes. Trends Parasitol. 2016, 32, 470–480.
- Sweeney, T.; Hanrahan, J.P.; Ryan, M.T.; Good, B. Immunogenomics of gastrointestinal nematode infection in ruminants—Breeding for resistance to produce food sustainably and safely. Parasite Immunol. 2016, 38, 569–586.
- Hayward, A.D. Causes and consequences of intra- and inter-host heterogeneity in defense against nematodes. Parasite Immunol. 2013, 35, 362–373.
- Ruiz-López, M.J.; Monello, R.J.; Gompper, M.E.; Eggert, L.S. The effect and relative importance of neutral genetic diversity for predicting parasitism varies across parasite taxa. PLoS ONE 2012, 9, e45404.
- Saddiqi, H.A.; Jabbar, A.; Sarwar, M.; Iqbal, Z.; Muhammad, G.; Nisa, M.; Shahzad, A. Small ruminant resistance against gastrointestinal nematodes: A case of Haemonchus contortus. Parasitol. Res. 2011, 109, 1483–1500.
- Acevedo-Whitehouse, K.; Gulland, F.; Greig, D.; Amos, W. Disease susceptibility in California sea lions. Nature 2003, 422, 35.
- Cassinello, J.; Gomendio, M.; Roldan, E.R.S. Relationship between coefficient of inbreeding and parasite burden in endangered gazelles. Conserv. Biol. 2001, 15, 1171–1174.
- Coltman, D.W.; Pilkington, J.G.; Smith, J.A.; Pemberton, J.M. Parasite-mediated selection against inbred Soay sheep in a free-living, island population. Evolution 1999, 53, 1259–1267.
- Hansson, B.; Westerberg, L. On the correlation between heterozygosity and fitness in natural populations. Mol. Ecol. 2002, 11, 2467–2474.
- Bateson, Z.W.; Hammerly, S.C.; Johnson, J.A.; Morrow, M.E.; Whittingham, L.A.; Dunn, P.O. Specific alleles at immune genes, rather than genome-wide heterozygosity, are related to immunity and survival in the critically endangered Attwater’s prairie chicken. Mol. Ecol. 2016, 25, 4730–4744.
- Brambilla, A.; Biebach, I.; Bassano, B.; Bogliani, G.; von Hardenberg, A. Direct and indirect causal effects of heterozygosity on fitness-related traits in Alpine ibex. Proc. R. Soc. B Biol. Sci. 2015, 82, 20141873.
- Janeway, C.A.; Travers, P.; Walport, M.; Shlomchik, M.J. The major histocompatibility complex and its functions. In The Immune System in Health and Disease, 5th ed.; Janeway, C., Ed.; Garland Science: New York, NY, USA, 2001.
- Aguilar, A.; Roemer, G.; Debenham, S.; Binns, M.; Garcelon, D.; Waine, R.K. High MHC diversity maintained by balancing selection in an otherwise genetically monomorphic mammal. Proc. Natl. Scad. Sci. USA 2004, 101, 3490–3494.
- Hughes, A.L.; Hughes, M.K.; Howell, C.Y.; Nei, M. Natural selection at the class II major histocompatibility complex loci of mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1994, 345, 359–367.
- Hedrick, P.W.; Thomson, G. Evidence for balancing selection at HLA. Genetics 1983, 104, 449–456.
- Spurgin, L.G.; Richardson, D.S. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. R. Soc. B Biol. Sci. 2010, 277, 979–988.
- Acevedo-Whitehouse, K.; Cunningham, A.A. Is MHC enough for understanding wildlife immunogenetics? Trends Ecol. Evol. 2006, 21, 433–438.
- Bernatchez, L.; Landry, C. MHC studies in nonmodel vertebrates: What have we learned about natural selection in 15 years? J. Evol. Biol. 2003, 16, 363–377.
- Turner, A.K.; Begon, M.; Jackson, J.A.; Paterson, S. Evidence for selection at cytokine loci in a natural population of field voles (Microtus agrestis). Mol. Ecol. 2012, 7, 1632–1646.
- Robinson, S.J.; Samuel, M.D.; Johnson, C.J.; Adams, M.; McKenzie, D.I. Emerging prion disease drives host selection in a wildlife population. Ecol. Appl. 2012, 22, 1050–1059.
- Johnson, C.; Johnson, J.; Vanderloo, J.P.; Keane, D.; Aiken, J.M.; McKenzie, D. Prion protein polymorphisms in white-tailed deer influence susceptibility to chronic wasting disease. J. Gen. Virol. 2006, 87, 2109–2114.
- White, P.S.; Choi, A.; Pandey, R.; Menezes, A.; Penley, M.; Gibson, A.K.; de Roode, J.; Morran, L. Host heterogeneity mitigates virulence evolution. Biol. Lett. 2020, 16, 20100744.
- Morley, D.; Broniewski, J.M.; Westra, E.R.; Buckling, A.; van Houte, S. Host diversity limits the evolution of parasite local adaptation. Mol. Ecol. 2017, 26, 1756–1763.
- Altizer, S.; Harvell, D.; Friedle, E. Rapid evolutionary dynamics and disease threats to biodiversity. Trends Ecol. Evol. 2003, 18, 589–596.
- Regoes, R.R.; Nowak, M.A.; Bonhoeffer, S. Evolution of virulence in a heterogeneous host population. Evolution 2000, 54, 64–71.
- Schmid-Hempel, P. Parasites in Social Insects; Princeton University Press: Princeton, NJ, USA, 1998.
- Anderson, R.M.; May, R.M. The invasion, persistence and spread of infectious diseases within animal and plant communities. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986, 314, 533–570.
- Ekroth, A.K.E.; Rafaluk-Mohr, C.; King, K.C. Host genetic diversity limits parasite success beyond agricultural systems: A meta-analysis. Proc. R. Soc. B Biol. Sci. 2019, 286, 20191811.
- van Houte, S.; Ekroth, A.K.; Broniewski, J.M.; Chabas, H.; Ashby, B.; Bondy-Denomy, J.; Gandon, S.; Boots, M.; Paterson, S.; Buckling, A.; et al. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 2016, 532, 385–388.
- King, K.C.; Lively, C.M. Does genetic diversity limit disease spread in natural host populations? Heredity 2012, 109, 199–203.
- Campbell, G.; Noble, L.R.; Rollinson, D.; Southgate, V.R.; Webster, J.P.; Jones, C.S. Low genetic diversity in a snail intermediate host (Biomphalaria pfeifferi Krass, 1848) and schistosomiasis transmission in the Senegal River Basin. Mol. Ecol. 2010, 19, 241–256.
- Bani, L.; Orioli, V.; Pisa, G.; Dondina, O.; Fagiani, S.; Fabbri, E.; Randi, E.; Mortelliti, A.; Sozio, G. Landscape determinants of genetic differentiation, inbreeding and genetic drift in the hazel dormouse (Muscardinus avellanarius). Conserv. Genet. 2017, 19, 283–296.
- Gubili, C.; Mariani, S.; Weckworth, B.V.; Galpern, P.; McDevitt, A.D.; Hebblewhite, M.; Nickel, B.; Musiani, M. Environmental and anthropogenic drivers of connectivity patterns: A basis for prioritizing conservation efforts for threatened populations. Evol. Appl. 2017, 10, 199–211.
- Charlesworth, B. Effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 2009, 10, 195–205.
- Epps, C.W.; Wehausen, J.D.; Bleich, V.C.; Torres, S.G.; Brashares, J.S. Optimizing dispersal and corridor models using landscape genetics. J. Appl. Ecol. 2007, 44, 714–724.
- Lawson Handley, J.L.; Perrin, N. Advances in our understanding of mammalian sex-biased dispersal. Mol. Ecol. 2007, 16, 1559–1578.
- Briton, J.; Nurthen, R.K.; Briscoe, D.A.; Frankham, R. Modelling problems in conservation genetics using Drosophila: Consequences of harem. Biol. Conserv. 1994, 69, 267–275.
- Slatkin, M. Gene flow and the geographic structure of natural populations. Science 1987, 236, 787–792.
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Scad. Sci. USA 1973, 70, 3321–3323.
- Wright, S. Size of population and breeding structure in relation to evolution. Science 1938, 87, 1417–1429.
- Arauco-Shapiro, G.; Schmacher, K.I.; Boersma, D.; Bouzat, J.L. The role of demographic history and selection in shaping genetic diversity of the Galápagos penguin (Spheniscus mendiculus). PLoS ONE 2020, 15, e0226439.
- Bouzat, J.L. Conservation genetics of population bottlenecks: The role of change, selection, and history. Conserv. Genet. 2010, 11, 463–478.
- Hedrick, P.W. Genetic of Populations; Jones & Bartlett Publishers: Sudbury, MA, USA, 2005.
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2002.
- Hedrick, P.W.; García-Dorado, A. Understanding inbreeding depression, purging, and genetic rescue. Trends Ecol. Evol. 2016, 31, 940–952.
- Groombridge, J.J.; Jones, C.G.; Bruford, M.W.; Nichols, R.A. ‘Ghost’ alleles of the Mauritius kestrel. Nature 2000, 403, 616.
- Bonnell, M.L.; Selander, R.L. Elephant seals: Genetic variation and near extinction. Science 1974, 134, 908–909.
- Hamilton, W.D.; Axelrod, R.; Tanese, R. Sexual reproduction as an adaptation to resist parasites (a review). Proc. Natl. Acad. Sci. USA 1990, 87, 3566–3573.
- Råberg, L.; Sim, D.; Read, A.F. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science 2007, 318, 812–814.
- Bishop, S.C.; Stear, M.J. Modeling of host genetics and resistance to infectious diseases: Understanding and controlling nematode infections. Vet. Parasitol. 2003, 115, 147–166.
- Blanchong, J.A.; Robinson, S.J.; Samuel, M.D.; Foster, J.T. Application of genetics and genomics to wildlife epidemiology. J. Wildl. Manage. 2016, 80, 593–608.
- Martinez-Padilla, J.; Vergara, P.; Mougeot, F.; Redpath, S.M. Parasitized mates increase infection risk for partners. Am. Nat. 2012, 179, 811–820.
- Arakawa, H.; Cruz, S.; Deak, T. From models to mechanisms: Odorant communication as a key determinant of social behavior in rodents during illness-associated states. Neurosci. Biobehav. Rev. 2011, 35, 1916–1928.
- Hillgarth, N. Ectoparasite transfer during matins in ring-necked pheasants Phasianus colchicus. J. Avian Biol. 1996, 27, 260–262.
- Clutton-Brock, T.H. The Evolution of Parental Care; Princeton University Press: Princeton, NJ, USA, 1991.
- Trivers, R.L. Parental investment and sexual selection. In Sexual Selection and the Descent of Man: 1871–1971; Campbell, B., Ed.; Aldine Press: Chicago, IL, USA, 1972; pp. 136–179.
- Setchell, J.M.; Charpentier, M.J.E.; Abbot, K.M.; Wickings, E.J.; Knapp, L.A. Opposites attract: MHC-associated mate choice in a polygynous primate. J. Evol. Biol. 2010, 23, 136–148.
- Penn, D.J. The scent of genetic compatibility: Sexual selection and the major histocompatibility complex. Ethology 2002, 108, 1–21.
- Penn, D.J.; Potts, W.K. The evolution of mating preferences and major histocompatibility complex genes. Am. Nat. 1999, 153, 145–164.
- Campbell, L.J.; Head, M.L.; Wilfert, L.; Griffiths, A.G.F. An ecological role for assortative mating under infection? Conserv. Genet. 2017, 18, 983–994.
- Teacher, A.G.F.; Garner, T.W.J.; Nichols, R.A. Population genetic patterns suggest a behavioural change in wild common frogs (Rana temporaria) following disease outbreaks (Ranavirus). Mol. Ecol. 2009, 18, 3163–3172.
- Bruniche-Olsen, A.; Burridge, C.P.; Austin, J.J.; Jones, M.E. Disease induced changes in gene flow patterns among Tasmanian devil populations. Biol. Conserv. 2013, 165, 69–78.
- Lachish, S.; McCallum, H.; Jones, M. Demography, disease and the devil: Life-history changes in a disease-affected population of Tasmanian devils (Sarcophilus harrisii). J. Anim. Ecol. 2009, 78, 427–436.
- Serieys, L.E.K.; Lea, A.; Pollinger, J.P.; Riley, S.P.D.; Wayne, R.K. Disease and freeways drive genetic change in urban bobcat populations. Evol. Appl. 2015, 8, 75–92.
- Van Valen, L. A new evolutionary law. Evol. Theory 1973, 1, 1–30.
- Gómez, P.; Ashby, B.; Buckling, A. Population mixing promotes arms race host-parasite coevolution. Proc. R. Soc. B Biol. Sci. 2015, 282, 20142297.
- Thornhill, R.; Fincher, C.L. The parasite-driven-wedge model of parapatric speciation. J. Zool. 2013, 291, 23–33.
- Fincher, C.L.; Thornhill, R. A parasite-driven wedge: Infectious diseases may explain language and other biodiversity. Oikos 2008, 117, 1289–1297.
- Han, B.A.; Kramer, A.M.; Drake, J.M. Global patterns of zoonotic disease in mammals. Trends Parasitol. 2016, 32, 565–577.
- Cleaveland, S.; Laurenson, M.K.; Taylor, L.H. Diseases of humans and their domestic mammals: Pathogen characteristics, host range and the risk of emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 991–999.
- Lloyd-Smith, J.O.; George, D.; Pepin, K.M.; Pitzer, V.E.; Pulliam, J.R.C.; Dobson, A.P.; Hudson, P.J.; Grenfell, B.T. Epidemic dynamics at the human-animal interface. Science 2009, 326, 1362–1367.
- Wolfe, N.D.; Dunavan, C.P.; Diamond, J. Origins of major human infectious diseases. Nature 2007, 447, 279–283.
- Dehove, A.; Commault, J.; Petitclerc, M.; Teissier, M.; Macé, J. Economic analysis and costing of animal health: A literature review of methods and importance. Rev. Sci. Tech. 2012, 31, 605–617.
- Williams, E.S.; Yuill, T.; Artois, M.; Fischer, J.; Haigh, S.A. Emerging infectious diseases in wildlife. Rev. Sci. Tech. 2002, 21, 39–157.
- Daszak, P.; Cunningham, A.A.A.; Hyatt, A.D. Emerging infectious diseases of wildlife—Threats to biodiversity and Human health. Science 2000, 287, 443–449.
- Ripple, W.J. Collapse of the world’s largest herbivores. Sci. Adv. 2015, 1, e1400103.
- Linnell, J.D.C.; Zachos, F.E. Status and distribution patterns of European ungulates: Genetics, population history and conservation. In Ungulate Management in Europe: Problems and Practices; Putman, R., Apollonio, M., Andersen, R., Eds.; Cambridge University Press: Cambridge, UK, 2011; pp. 12–53.
- Sæther, B.-E.; Engen, S.; Solberg, E.J. Effective size of harvested ungulate populations. Anim. Conserv. 2009, 12, 488–495.
- Coltman, D.W. Molecular ecological approaches to studying the evolutionary impact of selective harvesting in wildlife. Mol. Ecol. 2008, 17, 221–235.
- Queirós, J.; Vicente, J.; Alves, P.C.; de la Fuente, J.; Gortazar, C. Tuberculosis, genetic diversity and fitness in the red deer, Cervus elaphus. Infect. Genet. Evol. 2016, 43, 203–212.
- Kotzé, A.; Smith, R.M.; Moodley, Y.; Luikart, G.; Birss, C.; Van Wyk, A.M.; Dalton, D.L. Lessons for conservation management: Monitoring temporal changes in genetic diversity of cape mountain zebra (Equus zebra zebra). PLoS ONE 2019, 14, e0220331.
- Frantz, A.C.; Zachos, F.E.; Bertouille, S.; Eloy, M.C.; Colyn, M.; Flamand, M.C. Using genetic tolos to estimate the prevalence of non-native red deer (Cervus elaphus) in a western European population. Ecol. Evol. 2017, 7, 7650–7660.
- Strucken, E.M.; Lee, S.H.; Jang, G.W.; Porto-Neto, L.R.; Gondro, C. Towards breed formation by island model divergence in Korean cattle. BMC Evol. Biol. 2015, 15, 284.
- Queirós, J.; Vicente, J.; Boadella, M.; Gortázar, C.; Alves, P.C. The impact of management practices and past demographic history on the genetic diversity of red deer (Cervus elaphus): An assessment of population and individual fitness. Biol. J. Linn. Soc. 2014, 111, 209–223.
- Smitz, N.; Corneli, D.; Chardonnet, P.; Caron, A.; de Garine-Wichatitsky, M.; Jori, F.; Mouton, A.; Latinne, A.; Pigneur, L.M.; Melletti, M. Genetic structure of fragmented southern populations of African Cape buffalo (Syncerus caffer caffer). BMC Evol. Biol. 2014, 14, 203.
- Wan, Q.H.; Zhang, P.; Ni, X.W.; Wu, X.W.; Chen, Y.Y.; Kuang, Y.Y.; Ge, Y.F.; Fang, S.G. A novel HURRAH protocol reveals high numbers of monomorphic MHC class II loci and tow asymmetric multi-locus haplotypes in the Pere David’s deer. PLoS ONE 2011, 6, e14518.
- Fernández-de-Mera, I.G.; Vicente, J.; Pérez de la Lastra, J.M.; Mangold, A.J.; Naranjo, V.; Fierro, Y.; de la Fuente, J.; Gortázar, C. Reduced major histocompatibility complex class II polymorphism in a hunter-managed isolated Iberian red deer population. J. Zool. 2009, 277, 157–170.
- Wojcik, J.M.; Kawalko, A.; Tokarska, M.; Jaarola, M.; Vallenback, P.; Pertodi, C. Post-bottleneck mtDNA diversity in a free-living population of European bison: Implications for conservation. J. Zool. 2009, 277, 81–87.
- Wilson, G.A.; Nishi, J.S.; Elkin, B.T.; Strobeck, C. Effects of a recent founding event and intrinsic population dynamics on genetic diversity in an ungulate population. Conserv. Genet. 2005, 6, 905–916.
- Hedrick, P.W.; Parker, K.M.; Gutiérrez-Espeleta, G.A.; Rattink, A.; Lievers, K. Major histocompatibility complex variation in the Arabian oryx. Evolution 2000, 54, 2145–2151.
- Kumar, D.R.; Devadasan, M.J.; Surya, T.; Vineeth, M.R.; Choudhary, A.; Sivalingam, J.; Kataria, R.S.; Niranjan, S.K.; Tantia, M.S.; Verma, A. Genomic diversity and selection sweeps identified in Indian swamp buffaloes reveals it’s uniqueness with riverine buffaloes. Genomics 2020, 112, 2385–2392.
- McDonald, D.B.; Hobson, E.A. Edge weight variance: Population genetic metrics for social network analysis. Anim. Behav. 2018, 136, 239–250.
- Janova, E.; Futas, J.; Klumplerova, M.; Putnova, L.; Vrtkva, I.; Vyskocil, M.; Frolkova, P.; Horin, P. Genetic diversity and conservation in a small endangered horse population. J. Appl. Genet. 2013, 54, 285–292.
- Larison, B.; Kaelin, C.B.; Harrigan, R.; Henegar, C.; Rubenstein, D.I.; Kamath, P.; Aschenborn, O.; Smith, T.B.; Barsh, G.S. Population structure, inbreeding and stripe pattern abnormalities in plains zebras. Mol. Ecol. 2021, 30, 379–390.
- Coetzer, W.G.; Grobler, J.P. Genetic variation among different springbok (Antidorcas marsupialis) colour variants. Mol. Ecol. 2019, 99, 42–53.
- Sasidharan, S.P.; Ludwig, A.; Harper, C.; Moodley, Y.; Bertschinger, H.J.; Guthrie, A.J. Comparative genetics of sarcoid tumour-affected and non-affected mountain zebra (Equus zebra) populations. S. Afr. J. Wildl. Res. 2011, 41, 36–49.
- Pérez-González, J.; Carranza, J.; Torres-Porras, J.; Fernández-García, J.L. Low heterozygosity at microsatellite markers in Iberian red deer with small antlers. J. Hered. 2010, 101, 553–561.
- Marais, H.J.; Nel, P.; Bertschinger, H.J.; Schoeman, J.P.; Zimmerman, D. Prevalence and body distribution of sarcoids in South African Cape mountain zebra (Equus zebra zebra). J. S. Afr. Vet. Assoc. 2007, 78, 145–148.
- Zachos, F.E.; Althoff, C.; von Steynitz, Y.; Eckert, I.; Hartl, G.B. Genetic analysis of an isolated red deer (Cervus elaphus) population showing signs of inbreeding depression. Eur. J. Wildl. Res. 2007, 53, 61–67.
- Da Silva, A.; Gaillard, J.M.; Yoccoz, N.G.; Hewison, A.J.M.; Galan, M.; Coulson, T.; Allaine, D.; Vial, L.; Delorme, D.; Van Laere, G.; et al. Heterozygosity-fitness correlations revealed by neutral and candidate gene markers in roe deer from a long-term study. Evolution 2009, 63, 403–417.
- Kaeuffer, R.; Reale, D.; Pontier, D.; Chapuis, J.L.; Coltman, D.W. Local effects of inbreeding on embryo number and consequences for genetic diversity in Kerguelen mouflon. Biol. Lett. 2008, 4, 504–507.
- Latch, E.K.; Amann, R.P.; Jacobson, J.P.; Rhodes, O.E. Competing hypotheses for the etiology of cryptorchidism in sitka black-tailed deer: An evaluation of evolutionary alternatives. Anim. Conserv. 2008, 11, 234–246.
- Hermoso de Mendoza, J.; Parra, A.; Tato, A.; Alonso, J.M.; Rey, J.M.; Peña, A.; García-Sánchez, A.; Larrasa, J.; Teixido, J.; Manzano, G.; et al. Bovine tuberculosis in wild boar (Sus scrofa), red deer (Cervus elaphus) and cattle (Bos taurus) in a Mediterranean ecosystem (1992–2004). Prev. Vet. Med. 2006, 74, 239–247.
- Barasona, J.A.; Gortázar, C.; De La Fuente, J.; Vicente, J. Host Richness Increases Tuberculosis Disease Risk in Game-Managed Areas. Microorganisms 2019, 7, 182.
- Queirós, J.; Alves, P.C.; Vicente, J.; Gortázar, C.; de la Fuente, J. Genome-wide associations identify novel candidate loci associated with genetic susceptibility to tuberculosis in wild boar. Sci. Rep. 2018, 8, 1–12.
- Acevedo-Whitehouse, K.; Vicente, J.; Gortazar, C.; Höflem, U.; Fernández-de-Mera, I.; Amos, W. Genetic resistance to bovine tuberculosis in the Iberian wild boar. Mol. Ecol. 2005, 14, 3209–3217.
- Galarza, J.A.; Sánchez-Fernández, B.; Fandos, P.; Soriguer, R. Intensive management and natural genetic variation in red deer (Cervus elaphus). J. Hered. 2017, 108, 496–504.
- Galarza, J.; Sánchez-Fernández, B.; Fandos, P.; Soriguer, R. The genetic landscape of the Iberian red deer (Cervus elaphus hispanicus) after 30 years of big-game hunting in southern Spain. J. Wildl. Manage. Wildl. Monogr. 2015, 79, 500–504.
- Herrero-Medrano, J.M.; Megens, H.; Groenen, M.A.; Ramis, G.; Bosse, M.; Pérez-Enciso, M.; Crooijmans, R.P.M.A. Conservation genomic analysis of domestic and wild pig populations from the Iberian Peninsula. BMC Genet. 2013, 14, 106.
- Pérez-González, J.; Frantz, A.C.; Torres-Porras, J.; Castillo, L.; Carranza, J. Population structure, habitat features and genetic structure of managed red deer populations. Eur. J. Wildl. Res. 2012, 58, 933–943.
- Martínez, J.G.; Carranza, J.; Fernández-García, J.L.; Sánchez-Prieto, C.B. Genetic variation of red deer populations under hunting exploitation in Southwestern Spain. J. Wildl. Manage. 2002, 66, 1273–1282.
- Vicente, J.; Hofle, U.; Garrido, J.M.; Fernández-de-Mera, I.G.; Acevedo, P.; Juste, R.; Barral, M.; Gortazar, C. Risk factors associated with the prevalence of tuberculosis-like lesions in fenced wild boar and red deer in south central Spain. Vet. Res. 2007, 38, 451–464.
- Torres-Porras, J.; Carranza, J.; Pérez-González, J.; Mateos, C.; Alarcos, S. The tragedy of the commons: Unsustainable population structure of Iberian red deer in hunting estates. Eur. J. Wildl. Res. 2014, 60, 351–357.
- Pérez-González, J.; Carranza, J. Female-biased dispersal under conditions of low male mating competition in a polygynous mammal. Mol. Ecol. 2009, 18, 4617–4630.
- Pérez-González, J.; Costa, V.; Santos, P.; Slate, J.; Carranza, J.; Fernández-Llario, P.; Zsolnai, A.; Monteiro, N.M.; Anton, I.; Buzgó, J.; et al. Males and females contribute unequally to offspring genetic diversity in the polygynandrous mating system of wild boar. PLoS ONE 2014, 9, e115394.
- Carranza, J.; Pérez-González, J.; Mateos, C.; Fernández-García, J.L. Parents’ genetic dissimilarity and offspring sex in a polygynous mammal. Mol. Ecol. 2009, 18, 4964–4973.
- Pérez-González, J.; Mateos, C.; Carranza, J. Polygyny can increase rather than decrease genetic diversity contributed by males relative to females: Evidence from red deer. Mol. Ecol. 2009, 18, 1591–1600.
- Whiteley, A.R.; Fitzpatrick, S.W.; Funk, W.C.; Tallmon, D.A. Genetic rescue to the rescue. Trends Ecol. Evol. 2015, 30, 42–49.
- Bell, D.A.; Robinson, Z.L.; Funk, W.C.; Fitzpatrick, S.W.; Allendorf, F.W.; Talmon, D.A.; Whiteley, A.R. The exciting potential and remaining uncertainties of genetic rescue. Trends Ecol. Evol. 2019, 34, 1070–1079.
- Harris, K.; Zhang, Y.; Nielsen, R. Genetic rescue and the maintenance of native ancestry. Conserv. Genet. 2019, 20, 1–5.
- Carranza, J.; Martínez, J.G.; Sánchez-Prieto, C.; Fernández-García, J.L.; Sánchez-Fernández, B.; Álvarez-Álvarez, R.; Valencia, J.; Alarcos, S. Game species: Extinctions hidden by census numbers. Anim. Biodivers. Conserv. 2003, 26, 81–84.
- Frankham, R.; Ballou, J.D.; Eldridge, M.D.; Lacy, R.C.; Ralls, K.; Dudash, M.R.; Fenster, C.B. Predicting the probability of outbreeding depression. Conserv. Biol. 2011, 25, 465–475.
- Hedrick, P.W.; Fredrickson, R. Genetic rescue guidelines with examples from Mexican wolves and Florida panthers. Conserv. Genet. 2010, 11, 615–626.
- Pérez-Espona, S.; Pérez-Barberia, F.J.; McLeod, J.E.; Jiggins, C.D.; Gordon, I.J.; Pemberton, J.M. Landscape features affect gene flow of Scottish Highland red deer (Cervus elaphus). Mol. Ecol. 2008, 17, 981–996.
- Meagher, S.; Penn, D.J.; Potts, W.K. Male-male competition magnifies inbreeding depression in wild house mice. Proc. Natl. Scad. Sci. USA 2000, 97, 3324–3329.
- Pusey, A.; Wolf, M. Inbreeding avoidance in animals. Trends Ecol. Evol. 1996, 11, 201–206.
- Pearse, D.E.; Anderson, E.C. Multiple paternity increases effective population size. Mol. Ecol. 2009, 18, 3124–3127.
- Sugg, D.W.; Chesser, R.K. Effective population sizes with multiple paternity. Genetics 1994, 137, 1147–1155.
