Kinetoplastid parasites are responsible for high mortality and morbidity in (sub)tropical regions. In the development of new drugs, phenotypic screening already allowed the identification of promising new chemical entities with anti-kinetoplastid activity potential, but knowledge on their mode-of-action (MoA) is lacking due to the generally applied whole-cell based approach. However, identification of the drug target is highly beneficial to steer further hit finding, lead optimization and rational drug design. Multiple complementary ‘omics’ approaches have been successfully used to define the MoA or mode-of-resistance (MoR) of current reference drugs and some new anti-kinetoplastid compounds.
- Kinetoplastid
- Leishmaniasis
- Trypanosomiasis
- disease
- treatment
- drug
- drug target
- omics
- mode-of-action
- mode-of-resistance
1. Definition
Kinetoplastids are the causative agents of leishmaniasis, human African trypanosomiasis, and American trypanosomiasis. They are responsible for high mortality and morbidity in (sub)tropical regions. Adequate treatment options are limited and have several drawbacks, such as toxicity, need for parenteral administration, and occurrence of treatment failure and drug resistance. Therefore, there is an urgency for the development of new drugs. Phenotypic screening already allowed the identification of promising new chemical entities with anti-kinetoplastid activity potential, but knowledge on their mode-of-action (MoA) is lacking due to the generally applied whole-cell based approach. However, identification of the drug target is essential to steer further drug discovery and development. Multiple complementary techniques have indeed been used for MoA elucidation.
2. Introduction
Leishmaniasis, Chagas disease, and sleeping sickness are caused by kinetoplastid protozoan parasites and are responsible for high morbidity and mortality rates, especially in developing countries [1][2][3]. These diseases are characterized by severe clinical manifestations such as hepatosplenomegaly, cardiomyopathy, and neuropathology, all of which may lead to fatality if left untreated [4][5][6]. Current therapies are known to be less than adequate due to the suboptimal administration routes and long treatment duration, the occurrence of severe adverse effects, and the growing incidence of treatment failures [3][5][7]. It is evident that treatment options should be improved and that new drugs will be needed to sustain adequate disease control.
Target-based and phenotypic screening are two standard approaches adopted by the pharmaceutical industry to identify novel active chemical entities. So far, the former has not been very successful given the lack of fully validated targets and the limited knowledge on their molecular biology [3][4][8]. The few targets that have been suggested for Trypanosoma brucei and T. cruzi have only been partially characterized [8][9][10][11][12] and ornithine decarboxylase is currently the only fully validated target involved in the mode-of-action (MoA) of eflornithine [8][13]. Identification of novel drug ‘leads’ is generally still achieved by phenotypic cell-based screening [14][15][16][17] but with the disadvantage of lack of knowledge gained on the MoA. However, target elucidation remains pivotal for rational structure-based optimization of small molecules, which predicts adverse effects. Even though MoA studies are not essential for regulatory approval, they can strengthen successful drug development or redirect drug discovery [12][14][18][19][20]. Alternatively, knowledge on a drug’s mode-of-resistance (MoR) can also provide useful information for anti-kinetoplastid drug research [21].
As described in several reviews [18][19][22], multiple strategies can be used to identify the MoA of compounds. The present review specifically focuses on the different ‘omics’ approaches which in recent years have emerged as valuable techniques to evaluate the MoA and MoR of anti-kinetoplastid compounds (Figure 1).
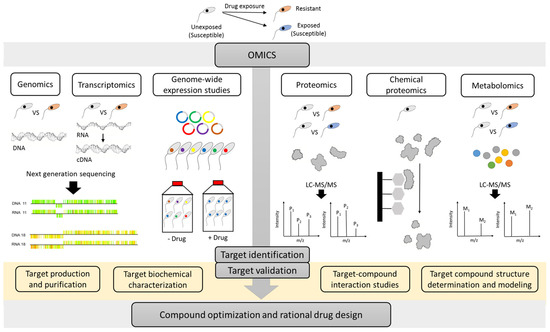
Figure 1. An overview of the multiple omics strategies that can be used to unravel the target of an anti-kinetoplastid compounds. Resistant parasites can be experimentally selected in the laboratory or obtained from the field. Comparative genomics, transcriptomics, proteomics/metabolomics approaches, genome-wide cosmid/RNAi libraries, and chemical proteomics can contribute complementary insights in the mode-of-action (MoA) or mode-of-resistance (MoR). Application of the various omics methodologies can enable the identification of drug targets, which may serve as starting point for additional hit finding, lead optimization and rational drug design. M: metabolite and P: protein. A graphical item in the figure about next generation sequencing was adopted from another publication [23].
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms8060950
References
- Barrett, M.P.; Burchmore, R.; Stich, A.; Lázzari, J.O.; Frasch, A.C.; Cazzulo, J.J.; Krishna, S; The trypanosomiases. Lancet 2003, 362, 1469–1480, .
- Hotez, P.J.; Fenwick, A.; Savioli, L.; Molyneux, D; Rescuing the bottom billion through control of neglected tropical diseases. Lancet 2009, 373, 1570–1575, .
- Rao, S.P.S.; Barrett, M.P.; Dranoff, G.; Faraday, C.J.; Gimpelewicz, C.R.; Hailu, A.; Jones, C.L.; Kelly, J.M.; Lazdins-Helds, J.K.; Mäser, P.; et al. Drug Discovery for Kinetoplastid Diseases: Future Directions. ACS Infect. Dis. 2018, 5, 152–157, .
- Bilal Zulfiqar; Todd B. Shelper; Vicky M. Avery; Leishmaniasis drug discovery: recent progress and challenges in assay development. Drug Discovery Today 2017, 22, 1516-1531, 10.1016/j.drudis.2017.06.004.
- Perez-Molina, J.A.; Molina, I; Chagas Disease. Lancet 2018, 391, 82-94, .
- Büscher, P.; Cecchi, G.; Jamonneau, V.; Priotto, G; Human African trypanosomiasis. Lancet 2017, 390, 2397–2409, .
- Catriona H. Baker; Susan C. Welburn; The Long Wait for a New Drug for Human African Trypanosomiasis. Trends in Parasitology 2018, 34, 818-827, 10.1016/j.pt.2018.08.006.
- Ian H. Gilbert; Drug Discovery for Neglected Diseases: Molecular Target-Based and Phenotypic Approaches. Journal of Medicinal Chemistry 2013, 56, 7719-7726, 10.1021/jm400362b.
- Adam R. Renslo; James H McKerrow; Drug discovery and development for neglected parasitic diseases. Nature Methods 2006, 2, 701-710, 10.1038/nchembio837.
- Cauê Benito Scarim; Daniela Hartmann Jornada; Rafael Consolin Chelucci; Letícia De Almeida; Jean Leandro Dos Santos; Man Chin Chung; Chung Man Chin; Current advances in drug discovery for Chagas disease. European Journal of Medicinal Chemistry 2018, 155, 824-838, 10.1016/j.ejmech.2018.06.040.
- Juan Felipe Osorio-Méndez; Ana María Cevallos; Discovery and Genetic Validation of Chemotherapeutic Targets for Chagas' Disease. Frontiers in Microbiology 2019, 8, 439, 10.3389/fcimb.2018.00439.
- Manu De Rycker; Beatriz Baragaña; Suzanne L. Duce; Ian H. Gilbert; Challenges and recent progress in drug discovery for tropical diseases. Nature 2018, 559, 498-506, 10.1038/s41586-018-0327-4.
- Nick V. Grishin; Andrei L. Osterman; Harold B. Brooks; Margaret A. Phillips; Elizabeth J. Goldsmith; X-ray Structure of Ornithine Decarboxylase fromTrypanosomabrucei: The Native Structure and the Structure in Complex with α-Difluoromethylornithine†,‡. Biochemistry 1999, 38, 15174-15184, 10.1021/bi9915115.
- Melissa L. Sykes; Vicky M. Avery; Approaches to Protozoan Drug Discovery: Phenotypic Screening. Journal of Medicinal Chemistry 2013, 56, 7727-7740, 10.1021/jm4004279.
- Fabiana Alves; Graeme Bilbe; Severine Blesson; Vishal Goyal; Séverine Monnerat; C.E. Mowbray; Gina Muthoni Ouattara; Bernard Pécoul; Suman Rijal; Joelle Rode; et al. Recent Development of Visceral Leishmaniasis Treatments: Successes, Pitfalls, and Perspectives. Clinical Microbiology Reviews 2018, 31, e00048-18, 10.1128/cmr.00048-18.
- Freitas-Junior, L.H.; Chatelain, E.; Kim, H.A.; Siqueira-Neto, J.L; Visceral leishmaniasis treatment: What do we have, what do we need and how to deliver it. Int. J. Parasitol. Drugs Drug Resist. 2012, 2, 11–19, .
- Michael Berninger; Ines Schmidt; Alicia Ponte-Sucre; Ulrike Holzgrabe; Novel lead compounds in pre-clinical development against African sleeping sickness. MedChemComm 2017, 8, 1872-1890, 10.1039/c7md00280g.
- Lindsay Tulloch; Stefanie Menzies; Ross P. Coron; Matthew D. Roberts; Gordon J. Florence; T. K. Smith; Direct and indirect approaches to identify drug modes of action. IUBMB Life 2017, 70, 9-22, 10.1002/iub.1697.
- Georg C. Terstappen; Christina Schlüpen; Roberto Raggiaschi; Giovanni Gaviraghi; Target deconvolution strategies in drug discovery. Nature Reviews Drug Discovery 2007, 6, 891-903, 10.1038/nrd2410.
- John G. Moffat; Fabien Vincent; Jonathan A. Lee; Jörg Eder; Marco Prunotto; Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nature Reviews Drug Discovery 2017, 16, 531-543, 10.1038/nrd.2017.111.
- Aya Hefnawy; Maya Berg; Jean-Claude Dujardin; Geraldine De Muylder; Exploiting Knowledge on Leishmania Drug Resistance to Support the Quest for New Drugs. Trends in Parasitology 2017, 33, 162-174, 10.1016/j.pt.2016.11.003.
- Marisa A. Azad; Gerard D. Wright; Determining the mode of action of bioactive compounds. Bioorganic & Medicinal Chemistry 2012, 20, 1929-1939, 10.1016/j.bmc.2011.10.088.
- Kumar, P.; Lodge, R.; Raymond, F.; Ritt, J.F.; Jalaguier, P.; Corbeil, J.; Tremblay, M.J; Gene expression modulation and the molecular mechanisms involved in Nelfinavir resistance in Leishmania donovani axenic amastigotes. Mol. Microbiol. 2013, 89, 565–582, .
