Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Medicine, Research & Experimental
Autophagy is a process of self-degradation that plays an important role in removing damaged proteins, organelles or cellular fragments from the cell.
- autophagy
- autophagy inhibitors
- autophagy activators
1. Introduction
Autophagy, directly translated as ‘self-eating’, is an evolutionary conservative process, found in all eukaryotic cells—from single-cell yeasts to much more complex multicellular mammalian organisms [1]. The introduction of the term ‘autophagy’ was proposed in February 1963 during the conference titled ‘Ciba Foundation Symposium on Lysosomes’ which took place in London [2]. This process participates in intracellular degradation of damaged or redundant proteins with a long half-life as well as other unnecessary cytoplasm components [3][4]. Autophagy provides an organism’s homeostasis and prevent it from redundant components accumulation inside the cell [5].
Moreover, this process is involved in surfactant formation or red blood cells ripening [3]. Following the Nomenclature Committee on Cell Death, in 2018 the term ‘autophagy-dependent cell death (ADCD)’ was introduced. ADCD is a type of regulated cell death in which functional autophagic markers such as increased degradation of autophagosomal substrates or LC3 (Light Chain protein 3) lipidization occurs [6]. Interestingly, unlike necrosis or apoptosis, autophagy-dependent cell death is not synonymous exclusively with cell death. Under stressful condition such as hypoxia, nutrient deficiency or chemotherapy, this process can become the strategy for cell survival [5]. ADCD occurs in all eukaryotic cells performing important functions, for example, it is an adaptation mechanism to stressful conditions, as it provides cells with a constant supply of nutrients essential for sustaining key life processes. Additionally, through the elimination of redundant cytoplasm components and the adjustment of the endoplasmic reticulum size, ADCD participates in maintaining the intracellular homeostasis. Furthermore, ADCD is involved in tissue-specific processes, such as erythrocyte ripening or intracellular surfactant formation [3] and also protects the organism from viruses or bacteria multiplication [7][8].
Autophagy-dependent cell death, through its selective and non-selective mechanisms of degradation of pathogens, organelles and various biomolecules (nucleic acids, lipids, carbohydrates and proteins) constitutes the main catabolic system of eukaryotic cells [9][10]. As one of the key elements in maintaining cell homeostasis and health, this process also plays an important role in tumor suppression or genome integrity [11].
2. Autophagy and Programmed Cell Death—Double-Edged Sword Relationship
Autophagy and apoptosis are regulated in the cell by different mechanisms. However, it happens that both processes overlap. Under the influence of stress, sequential or simultaneous activation of the apoptotic and autophagy pathways can occur in a cell. There are potential pathways of the relationship between apoptosis and autophagy: activation of autophagy and subsequent inhibition of apoptosis, activation of autophagy leading to activation of the apoptotic pathway, autophagy suppression and induction of apoptosis or activation of autophagy and apoptosis simultaneously, leading to cell death on apoptotic and autophagy-dependent pathway (Figure 1) [12][13][14]. In the former case, the cell activates autophagy in response to a stress signal. As a defense mechanism, autophagy leads to the removal of damaged fragments, preventing the activation of the apoptotic pathway. The second possibility is a situation in which the cell is no longer able to defend itself against the resulting damage, and the activated autophagy subsequently leads to activation of apoptosis and cell death. In the last case, a stress signal triggers both processes, resulting in cell death via two pathways [12]. Key factors connecting apoptosis and autophagy include, for example: p53, Bcl-2/Beclin 1, Atg proteins, p62 or caspases.
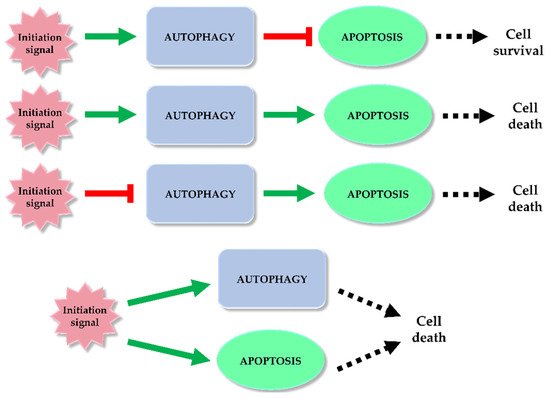
Figure 1. Autophagy and apoptosis relationship. Among the potential correlation pathways between autophagy and programmed cell death we can distinguish: activation of autophagy and apoptosis inhibition, activation of autophagy and activation of the apoptotic pathway, autophagy suppression and induction of apoptosis or simultaneous activation of autophagy and apoptosis leading to ADCD and apoptosis. The inhibitory effect of each process (red mark) and inducting effect (green mark) is indicated on the scheme.
2.1. p53 in Apoptosis and Autophagy
p53, a protein which binds specific DNA sequences, is involved in many cellular processes including repair of damaged DNA and induction of apoptosis. Due to its ability to regulate the cell cycle, p53 is called the guardian of the genome [15]. Activation of this factor can occur, for example, as a result of DNA damage, hypoxia, or nutritional stress [16][17][18]. p53 can affect both the extrinsic and intrinsic pathway of apoptosis. DNA damage causes mitochondrial translocation of p53. The protein promotes cytoplasmically localized Fas and TRAIL receptors, leading to induction of the extrinsic apoptotic pathway [19][20]. However, in the cell nucleus, p53 promotes many proapoptotic proteins, such as Bid, PUMA or Bax. In addition, it leads to inhibition of Bcl-2 expression, and both of these actions trigger the intrinsic apoptosis pathway [20].
The p53 protein is also involved in the regulation of autophagy. Based on their study, Crighton and co-authors found that genotoxic stress results in transcriptional activation of DRAM (Damage-Regulated Autophagy Modulator), a direct target gene of p53, which causes induction of autophagy. The DRAM signaling cascade promotes the fusion of autophagosomes and lysosomes, resulting in the formation of autolysosomes. This p53 target gene is an essential factor in the proper functioning of the apoptosis regulatory network and p53-dependent autophagy [21]. Furthermore, Tasdemir et al. demonstrated that cytoplasmically localized p53 through inactivation of AMPK and subsequent activation of the mTOR signaling pathway leads to inhibition of autophagy in the cell [22].
Scherz-Shouval and co-authors detected a relationship between autophagy and apoptosis processes. They revealed that under starvation conditions, p53 post-translationally inhibits the regulation of LC3 level, which leads to its accumulation in cells and decreases the rate of the autophagy process. The consequence is cell death by apoptosis [23].
2.2. Bcl-2/Beclin 1 in Apoptosis and Autophagy
Bcl-2, members of the B-cell lymphoma family of proteins, inhibits the release of cytochrome c from the mitochondrial interior, thereby playing a key role in the intrinsic apoptotic pathway [24]. Beclin 1 is a key element involved in autophagosome formation and is also an important component of the PI3K/Vps34 class III complex [25]. Bcl-2 binding to Beclin 1 leads to dissociation of Beclin 1 from PI3K class III, which results in inhibition of autophagy [26]. However, the occurrence of mutation in the BH3 receptor domain of Bcl-2 or Beclin 1 domain leads to dysfunction of Bcl-2/Beclin 1 complex, intensification of autophagy and promotion of cell survival [27][28].
Under nutrient-deficient conditions, autophagy is an essential element for cell survival. Activation of JNK1 (C-Jun N-terminal protein Kinase 1) and phosphorylation of residues involved in the Bcl-2′s regulatory loop lead to the destruction of the Bcl-2/Beclin 1 complex and consequently to initiation of autophagy [29]. Under standard conditions the phosphorylated Bcl-2 molecule binds to Bax, leading to inhibition of apoptosis. Due to the normal phosphorylation of Bcl-2, it is possible to maintain the integrity of the mitochondrial membrane, which in turn protects cells from death by the intrinsic apoptotic pathway. Sustaining the integrity of the mitochondrial membrane prevents the release of proapoptotic proteins from within the organelle into the cytoplasm [30]. However, in the situation of long-term nutrient deficiency, autophagy is not able to alleviate cellular damages. Intensification of Bcl-2 phosphorylation (hyperphosphorylation) promoted by JNK1 occurs [31]. This results in dissociation of the Bcl-2 molecule from Bax and apoptotic cell death. When the cell receives adequate amounts of nutrients, Bax/Bak and Beclin 1 bind to Bcl-XL or Bcl-2, leading to the inhibition of activation of both processes, apoptosis and autophagy [28][32].
2.3. Atg Proteins in Apoptosis and Autophagy
The level of autophagy-related proteins in a cell is regulated by the availability of growth factors and nutrients essential for proper cell functioning. Among Atg proteins we can distinguish the Atg12–Atg5 complex, which is important in both autophagy and apoptosis [33].
The Atg12–Atg5 complex, essential for autophagosome formation, also participates in the apoptotic pathway in an unconjugated form. Atg12 binding through a BH3-like motif to Bcl-2 and Mcl-1 (Myeloid Cell Leukemia 1) increases the intensity of the intrinsic apoptotic pathway. Interestingly, the anti-apoptotic properties of Mcl-1 can be inhibited in the cell as a result of abnormal Atg12 expression. Moreover, silencing Atg12 in an apoptotic cell will result in the inhibition of Bax induction and the arrest of cytochrome c release from the mitochondrion [34]. Cleaved by cell stress-activated cysteine proteases (caplains), Atg5 plays a significant role in the initiation of the intrinsic apoptosis pathway. As a consequence of cleavage, translocation of the N-terminal part of the Atg5 protein into the mitochondrion occurs. Inside the organelle, this fragment interacts with Bcl-XL allowing Atg-5 to be involved in the release of cytochrome c from the mitochondrion and indirectly participating in apoptosis promotion [35]. Taken together, Atg5 and Atg12 proteins may be involved in both autophagy and apoptosis, depending on the cellular conditions.
2.4. p62 in Apoptosis and Autophagy
p62, also known as SQSTM1, is a multi-domain adaptor protein that controls cell viability by regulating both autophagy and apoptosis [36]. By polymerizing with other p62 molecules, this protein has the ability to accumulate ubiquitin-tagged proteins. Aggregates of p62 (called p62 speckles), through their storage properties and ability to bind to the LC3 molecule, recognize, gather, and most importantly transport cargo to the autophagosomes [15]. p62, through its ability to activate caspase-8 on the autophagosome membrane also plays an important role in the induction of apoptosis. The autophagy-dependent mechanism of caspase-8 activation involves simultaneous induction of autophagy and activation of caspase-8. The autophagosomal membrane provides some kind of platform on which the caspase cascade leading to cell death by apoptosis is initiated. Depletion of Atg3 or Atg5 leads to the suppression of autophagosome formation, which in turn results in the inhibition of caspase-8 activation and subsequent suppression of apoptosis [37].
2.5. Caspases in Apoptosis and Autophagy
Caspases, enzymes belonging to the group of cysteine proteases, have been known to science for a long time. Their participation and the exact mechanism of action in the process of apoptosis have been widely studied and described in many scientific articles [38][39][40]. These enzymes are involved in both intrinsic and extrinsic pathways of apoptosis, acting as initiators (caspases-2, -8, -9 and -10) or effectors (3, -6 and -7) [41]. Caspases under standard conditions occurring in the form of inactive zymogenic precursors can be activated under the influence of various external or internal stimuli that initiate apoptosis. Activated enzymes may participate in the apoptotic pathway [42]. Despite the significant differences between the autophagy and apoptosis processes, the conducted studies indicate that caspases also affect the autophagy process. Oral and co-authors have shown that overexpression of caspase-8 leads to degradation of Atg3 protein and thus prevents its pro-autophagic activity [43]. Furthermore, Wirawan et al. showed that two key components of the autophagy-inducing complex (class III PI3K and Beclin 1) are direct substrates of caspases. It was observed that in response to different signals inducing the two apoptotic pathways, these enzymes cause cleavage of the complex components. Thus, the researchers confirmed that class III PI3K and Beclin 1 are substrates of caspases [44]. In contrast, Han and co-authors showed that caspase-9, by promoting Atg7-dependent LC3-II transformation, facilitates autophagosome formation. Moreover, the authors showed that depending on the cellular conditions, Atg7 can also form a complex with caspase-9 and directly inhibit the proapoptotic activity of the enzyme [45]. All of these studies indicate there is a mutual correlation between autophagy and apoptosis processes.
This entry is adapted from the peer-reviewed paper 10.3390/ijms22115804
References
- Levine, B.; Klionsky, D.J. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell 2004, 6, 463–477.
- Kroemer, G.; Galluzzi, L.; Vandenabeele, P.; Abrams, J.; Alnemri, E.S.; Baehrecke, E.H.; Blagosklonny, M.V.; El-Deiry, W.S.; Golstein, P.; Green, D.R.; et al. Classification of cell death: Recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009, 16, 3–11.
- Kim, R. Recent advances in understanding the cell death pathways activated by anticancer therapy. Cancer 2005, 103, 1551–1560.
- Polewska, J. Autophagy—molecular mechanism, apoptosis and cancer. Postepy Hig. Med. Dosw. 2012, 66, 921–936.
- Dereń-Wagemann, I.; Kiełbiński, M.; Kuliczkowski, K. Autofagia—Proces o dwóch obliczach. Acta Haematol. Pol. 2013, 44, 383–391.
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541.
- Klionsky, D.J. The molecular machinery of autophagy: Unanswered questions. J. Cell Sci. 2005, 118, 7–18.
- Yang, Y.-P.; Liang, Z.-Q.; Gu, Z.-L.; Qin, Z.-H. Molecular mechanism and regulation of autophagy. Acta Pharmacol. Sin. 2005, 26, 1421–1434.
- Rabinowitz, J.D.; White, E. Autophagy and metabolism. Science 2010, 330, 1344–1348.
- Eskelinen, E.-L.; Saftig, P. Autophagy: A lysosomal degradation pathway with a central role in health and disease. Biochim. Et Biophys. Acta (BBA) Mol. Cell Res. 2009, 1793, 664–673.
- Andrade-Tomaz, M.; de Souza, I.; Rocha, C.R.R.; Gomes, L.R. The role of chaperone-mediated autophagy in cell cycle control and its implications in cancer. Cells 2020, 9, 2140.
- Condello, M.; Pellegrini, E.; Caraglia, M.; Meschini, S. Targeting autophagy to overcome human diseases. Int. J. Mol. Sci. 2019, 20, 725.
- Chen, Q.; Kang, J.; Fu, C. The independence of and associations among apoptosis, autophagy, and necrosis. Signal. Transduct. Target. Ther. 2018, 3, 18.
- Mariño, G.; Niso-Santano, M.; Baehrecke, E.H.; Kroemer, G. Self-consumption: The interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2014, 15, 81–94.
- Hu, C.-A.A.; White, K.; Torres, S.; Ishak, M.-A.; Sillerud, L.; Miao, Y.; Liu, Z.; Wu, Z.; Sklar, L.; Berwick, M. Chapter 10—Apoptosis and autophagy: The yin–yang of homeostasis in cell death in cancer. In Autophagy: Cancer, Other Pathologies, Inflammation, Immunity, Infection, and Aging; Hayat, M.A., Ed.; Academic Press: Amsterdam, The Netherlands, 2015; pp. 161–181.
- Gaglia, G.; Lahav, G. Constant rate of p53 tetramerization in response to DNA damage controls the p53 response. Mol. Syst. Biol. 2014, 10, 753.
- Leszczynska, K.B.; Foskolou, I.P.; Abraham, A.G.; Anbalagan, S.; Tellier, C.; Haider, S.; Span, P.N.; O’Neill, E.E.; Buffa, F.M.; Hammond, E.M. Hypoxia-induced p53 modulates both apoptosis and radiosensitivity via AKT. J. Clin. Investig. 2015, 125, 2385–2398.
- Reid, M.A.; Wang, W.I.; Rosales, K.R.; Welliver, M.X.; Pan, M.; Kong, M. The B55α subunit of PP2A drives a p53-dependent metabolic adaptation to glutamine deprivation. Mol. Cell 2013, 50, 200–211.
- Li, M.; Gao, P.; Zhang, J. Crosstalk between autophagy and apoptosis: Potential and emerging therapeutic targets for cardiac diseases. Int. J. Mol. Sci. 2016, 17, 332.
- Fridman, J.S.; Lowe, S.W. Control of apoptosis by p53. Oncogene 2003, 22, 9030–9040.
- Crighton, D.; Wilkinson, S.; O’Prey, J.; Syed, N.; Smith, P.; Harrison, P.R.; Gasco, M.; Garrone, O.; Crook, T.; Ryan, K.M. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006, 126, 121–134.
- Tasdemir, E.; Maiuri, M.C.; Galluzzi, L.; Vitale, I.; Djavaheri-Mergny, M.; D’Amelio, M.; Criollo, A.; Morselli, E.; Zhu, C.; Harper, F.; et al. Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 2008, 10, 676–687.
- Scherz-Shouval, R.; Weidberg, H.; Gonen, C.; Wilder, S.; Elazar, Z.; Oren, M. p53-dependent regulation of autophagy protein LC3 supports cancer cell survival under prolonged starvation. Proc. Natl. Acad. Sci. USA 2010, 107, 18511–18516.
- Kilbride, S.M.; Prehn, J.H.M. Central roles of apoptotic proteins in mitochondrial function. Oncogene 2013, 32, 2703–2711.
- McKnight, N.C.; Yue, Z. Beclin 1, an essential component and master regulator of PI3K-III in health and disease. Curr. Pathobiol. Rep. 2013, 1, 231–238.
- Decuypere, J.-P.; Parys, J.B.; Bultynck, G. Regulation of the autophagic Bcl-2/Beclin 1 interaction. Cells 2012, 1, 284–312.
- Kang, R.; Zeh, H.J.; Lotze, M.T.; Tang, D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011, 18, 571–580.
- Maiuri, M.C.; Criollo, A.; Tasdemir, E.; Vicencio, J.M.; Tajeddine, N.; Hickman, J.A.; Geneste, O.; Kroemer, G. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-XL. Autophagy 2007, 3, 374–376.
- Wei, Y.; Pattingre, S.; Sinha, S.; Bassik, M.; Levine, B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell 2008, 30, 678–688.
- Ruvolo, P.P.; Deng, X.; May, W.S. Phosphorylation of Bcl2 and regulation of apoptosis. Leukemia 2001, 15, 515–522.
- Wei, Y.; Sinha, S.C.; Levine, B. Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy 2008, 4, 949–951.
- van Delft, M.F.; Huang, D.C.S. How the Bcl-2 family of proteins interact to regulate apoptosis. Cell Res. 2006, 16, 203–213.
- Cooper, K.F. Till death do us part: The marriage of autophagy and apoptosis. Oxidative Med. Cell. Longev. 2018, 2018, 4701275.
- Rubinstein, A.D.; Eisenstein, M.; Ber, Y.; Bialik, S.; Kimchi, A. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol. Cell 2011, 44, 698–709.
- Yousefi, S.; Perozzo, R.; Schmid, I.; Ziemiecki, A.; Schaffner, T.; Scapozza, L.; Brunner, T.; Simon, H.-U. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 2006, 8, 1124–1132.
- Huang, S.; Okamoto, K.; Yu, C.; Sinicrope, F.A. p62/sequestosome-1 up-regulation promotes ABT-263-induced caspase-8 aggregation/activation on the autophagosome *. J. Biol. Chem. 2013, 288, 33654–33666.
- Young, M.M.; Takahashi, Y.; Khan, O.; Park, S.; Hori, T.; Yun, J.; Sharma, A.K.; Amin, S.; Hu, C.-D.; Zhang, J.; et al. Autophagosomal membrane serves as platform for intracellular Death-inducing Signaling Complex (iDISC)-mediated caspase-8 activation and apoptosis *. J. Biol. Chem. 2012, 287, 12455–12468.
- Gornowicz, A.; Bielawska, A.; Szymanowski, W.; Gabryel-Porowska, H.; Czarnomysy, R.; Bielawski, K. Mechanism of anticancer action of novel berenil complex of platinum(II) combined with anti-MUC1 in MCF-7 breast cancer cells. Oncol. Lett. 2018, 15, 2340–2348.
- Pawłowska, N.; Gornowicz, A.; Bielawska, A.; Surażyński, A.; Szymanowska, A.; Czarnomysy, R.; Bielawski, K. The molecular mechanism of anticancer action of novel octahydropyrazino[2,1-a:5,4-a′]diisoquinoline derivatives in human gastric cancer cells. Investig. New Drugs 2018, 36, 970–984.
- Gornowicz, A.; Pawłowska, N.; Czajkowska, A.; Czarnomysy, R.; Bielawska, A.; Bielawski, K.; Michalak, O.; Staszewska-Krajewska, O.; Kałuża, Z. Biological evaluation of octahydropyrazin[2,1-a:5,4-a′]diisoquinoline derivatives as potent anticancer agents. Tumor Biol. 2017, 39, 1010428317701641.
- Wu, H.; Che, X.; Zheng, Q.; Wu, A.; Pan, K.; Shao, A.; Wu, Q.; Zhang, J.; Hong, Y. Caspases: A molecular switch node in the crosstalk between autophagy and apoptosis. Int. J. Biol. Sci. 2014, 10, 1072–1083.
- Parrish, A.B.; Freel, C.D.; Kornbluth, S. Cellular mechanisms controlling caspase activation and function. Cold Spring Harb. Perspect Biol. 2013, 5.
- Oral, O.; Oz-Arslan, D.; Itah, Z.; Naghavi, A.; Deveci, R.; Karacali, S.; Gozuacik, D. Cleavage of Atg3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis 2012, 17, 810–820.
- Wirawan, E.; Vande Walle, L.; Kersse, K.; Cornelis, S.; Claerhout, S.; Vanoverberghe, I.; Roelandt, R.; De Rycke, R.; Verspurten, J.; Declercq, W.; et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010, 1, e18.
- Han, J.; Hou, W.; Goldstein, L.A.; Stolz, D.B.; Watkins, S.C.; Rabinowich, H. A complex between Atg7 and caspase-9: A novel mechanism of cross-regulation between autophagy and apoptosis*. J. Biol. Chem. 2014, 289, 6485–6497.
This entry is offline, you can click here to edit this entry!
