Nowadays, biotechnology represents our best resource both for protecting crop yield and for a science-based increased sustainability in agriculture. Over the last decades, agricultural biotechnologies have made important progress based on the diffusion of new, fast and efficient technologies, offering a broad spectrum of options for understanding plant molecular mechanisms and breeding. This knowledge is accelerating the identification of key resistance traits to be rapidly and efficiently transferred and applied in crop breeding programs.
- crop disease resistance
- plant-microbe interaction
- molecular mechanisms in plant immunity
- sustainable agriculture
- Plant biotechnology
1. Introduction
Food availability and security challenge may be overcome by boosting crop yield, particularly that of cereals, and/or by reducing crop yield losses (20–40%) to pests and diseases, therefore diminishing further consequences for livelihoods, public health and the environment [1]. Moreover, effectiveness of long-term use of pesticides is impeded by different levels of resistance developed by phytopathogens [2]. Crop rotation, aiming to prevent the pathogen accumulation by alternating an incompatible host, together with the introduction of plant disease resistance genes (R genes) through specific breeding programs, represents alternative methods to combat yield losses to pests.
In this scenario, it would be very difficult, if not impossible, to succeed with conventional breeding, and the role of plant sciences and biotechnology becomes crucial for the future of humankind. Therefore, to find harmless control strategies for crop disease management, we need to exploit the plant innate immunity that, if timely activated, can efficiently contrast and restrict plant infection by microorganisms. In fact, although in nature plants face many types of biotic stresses caused by various organisms including fungi, viruses, bacteria, nematodes and insects, they generally resist most pathogens, and plant infection is usually the exception, not the rule [3].
Plants possess an innate ability to sense and recognize potential invading microorganisms and to mount successful defenses. Only pathogens with an evolved ability to evade recognition or suppress host defense mechanisms, or both, are successful. These biotic stress agents cause different kinds of diseases, infections, and damage to cultivated plants and significantly impact crop productivity [4].
2. Plant Biotechnology: From Random to Directed, Precise and Safe Mutagenesis
Over thousands years since 10,000 BP, humans have domesticated plants in an unconscious manner, selecting phenotypes with traits essential either for wide adaptation to different environments or improved agronomic performance. At the turn of 19th century, the introduction of Mendelian laws led to a scientific approach in crop breeding, thus representing the first revolution in the field of plant science (Figure 1).
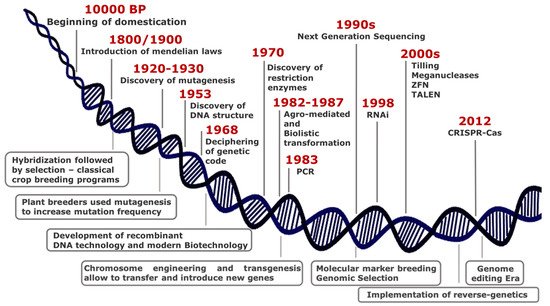
Although the most common way of generating genetic variability is to mate (cross) two or more parents that have contrasting genotypes, the selection of best resulting phenotypes fostered the development of monotypic crop fields, with consequent loss of biodiversity.
Genetic variability is the basis to discover new beneficial traits and results from mutations that have occurred in genomes, either naturally or induced.
Plant breeders have used mutagenesis intensively since 1950, and to date, the FAO/IAEA Mutant Varieties Database includes more than 3300 varieties that have been released worldwide for commercial use.
Some important achievements in plant sciences characterized the second half of the last century, among which the genetic engineering technology including chromosome engineering and transgenesis for gene transfer between species distantly related.
Genetic manipulation quickly proved to have a great potential in functional genomics contributing to unravel essential in plant physiology mechanisms. In few years, transgenesis
was widely adopted in plant breeding programs since it renders possible introgression of genes or any DNA sequence from other species and enables targeted editing of plant genome to increase genetic variability.
In the last decades, new breeding techniques (NBTs) are rapidly emerging from advances in genomic research and for application in crop traits improvement. They enable precise, targeted, and reliable changes in the genome and do not create multiple, unknown, unintended mutations, unlike chemical or radiation-induced mutagenesis.
Genome-editing methods produce defined mutants, thus becoming a potent tool in functional genomics and crop breeding. Zinc Finger Nucleases (ZFN) and Transcription Activator-Like Effector Nucleases (TALENs) were the dominant genome editing tools until the rise of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and Crispr associated protein (Cas).
For the first time ever, researchers and breeders can select and target any location in the genome by the use of a short synthetic guide RNA (sgRNA) along with an endonuclease enzyme (Cas9) [[5]]. The discovery of new Cas9 orthologs (Cpf1, Cas13) and the introduction of prime editing by fusing Cas9 to reverse transcriptase [6] enable to extend genome editing applications [7][8][9][10]. Such technology is applied in a wide range of applications spanning from gene silencing and gene insertions to base, RNA, and epigenome editing
For several genome-editing techniques, the resultant plants are free from foreign genes and would be indistinguishable both from plants generated by conventional breeding techniques and from naturally mutated plants.
Indeed, a recently published study of the European Commission regarding the status of new genomic techniques (NGT) under Union law identified limitations to the capacity of the legislation to keep pace with scientific developments, causing implementation challenges and legal uncertainties. It concluded that the applicable legislation is not fit for the purpose of some NGTs and their products and that it needs to be adapted to scientific and technological progress.
3. Increasing Disease-Resistance in Cereals by Implementing Plant Immunity Through Transgenesis
During evolutionary warfare with pathogens, plants have evolved sophisticated detection and inducible defense systems to properly defend themselves (Figure 2). Therefore, plants deploy hundreds of pattern recognition receptors (PRRs) in the cell plasma membrane, conceptually analogous to Toll-like receptors in animal cells [11], that can identify both non-self-molecules, referred to as pathogen-associated molecular patterns (PAMPs), and altered self-molecules or damage-associated molecular patterns (DAMPs) [12][13]. Ligand binding by its cognate receptor, belonging to the Receptor-Like Kinases (RLKs) or Receptor-Like Proteins (RLPs) classes, triggers the socalled PAMP/DAMP-triggered immunity (P/DTI).
A second level of the plant immune system involves plant resistance proteins able to recognize pathogen specific effectors (Avr proteins) and triggers plant defense mechanisms in a more robust way [14]. This kind of resistance is called effector-triggered immunity (ETI). Most resistance genes (R genes) encode proteins with unique domains that contain a conserved Nucleotide Binding Site called NBS. LRR (Leucin-Rich Repeat) is the second most important domain. NB-LRR receptors may recognize pathogen effectors delivered inside the cell to favor plant colonization [15].
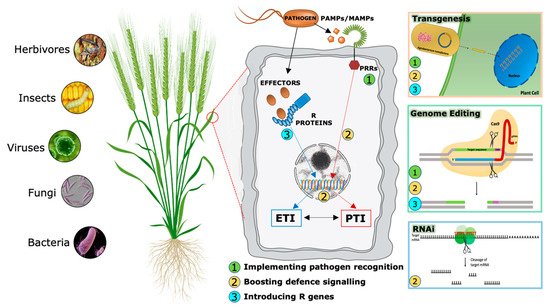
Traditionally, PTI and ETI have been considered to act sequentially but independently. However, recent accumulating evidence shows that the distinction between PAMPs and effectors, PRRs and R proteins, therefore between PTI and ETI, cannot strictly be maintained [16][17], suggesting an alternative model in which the two systems interact and share common elements but in which the cellular responses they evoke appear to be distinct. Analyses of specific mutants concluded that the activation of PTI is essential for ETI to function, while ETI can boost the efficiency of PTI and prolong the immune response duration.
Plant hormones, or phytohormones, are naturally occurring signaling compounds with diverse chemical properties. The activity of a given hormone depends on its biosynthesis, conjugation, transport, and degradation as well as hormone activation and inactivation [18][19]. Although all hormones regulate several processes independently, inducible defense responses are fine-tuned by very complex crosstalk among hormone signaling outputs [20][21][22]. Such a complex and multilayered plant immune system offers different levels on which researchers could act through biotechnological approaches in order to enhance or implement plant resistance (Table 1).
| Immunity Level of Intervention | Biotechnological Intervention | Gene | Species | Enhanced Resistance to | References |
|---|---|---|---|---|---|
| Pathogen sensing | Interspecies/interfamily transfer of known PRRs | AtEFR | Wheat | Pseudomonas syringae pv. oryzae | [23] |
| AtEFR | Rice | Xanthomonas oryzae pv. oryzae-derived elf18 | [24] | ||
| AtEFR | Rice | Acidovorax avenae subsp. avenae | [25] | ||
| OsXa21 | Rice | Xanthomonas oryzae pv. oryzae | [26] | ||
| TaRLK1 and TaRLK2 | Wheat | Blumeria graminis f. sp. tritici | [27] | ||
| HvLEMK1 | Barely, Wheat | Blumeria graminis f.sp. hordei; Blumeria graminis f. sp. tritici | [28] | ||
| HvLecRK-V | Wheat | Blumeria graminis f. sp. tritici | [29] | ||
| Production of chimeric receptor kinases and R genes | AtEFR-OsXa21 | Rice | Pseudomonas syringae pv. tomato; Agrobacterium tumefaciens; Xanthomonas oryzae pv. oryzae | [30][31] | |
| OsXa21-OsCEPiP | Rice | Magnaporthe oryzae | [32] | ||
| Effector detection | Deletion of effector binding sites | Os11N3/OsSWEET14 | Rice | Xanthomonas oryzae pv. oryzae | [33] |
| Addition of effector binding sites | OsXa27 | Rice | Xanthomonas oryzae pv. oryzae | [34] | |
| Immune signaling | Altered expression of signaling components | AtNPR1 | Rice | Broad-spectrum of pathogens | [35] |
| Altered expression of transcription factors | TaPIMP1 | Wheat | Bipolaris sorokiniana | [36] | |
| OsIPA1/OsSPL14 | Rice | Magnaporthe oryzae | [37] | ||
| R genes | Transfer of APR alleles | TaLr34 | Barely, Rice, Sorghum Maize, Durum wheat | Multiple biotrophic pathogens | [38][39][40][41][42] |
| TaLr67 | Barely | Multiple rusts and powdery mildew | [43] |
3.1. Pathogen Detection
Knowledge of the plant immune system offers the opportunity to develop new strategies of intervention at the pathogen perception level (Table 1). Increased or new recognition ability may be generated in different ways, for example by intra- and interspecies introduction of PRRs from other plants with novel recognition specificity [11][27][28][44][45][46]. In a recent study, the Arabidopsis thaliana EF-Tu (elongation factor thermo unstable) receptor, abbreviated as EFR, was transferred to monocot rice to confer resistance to two Xanthomonas oryzae pv. Oryzae isolates. Rice plants expressing such receptors were able to sense the bacterial ligand of EFR and to elicit an immune response. AtEFR was also expressed in wheat [23] driven by the rice actin promoter, and the plants showed enhanced induction of defense-related genes, callose deposition, and resistance against the cereal bacterial pathogen P. syringae pv. Oryzae. In another study, a lectin receptor-like kinase gene (LecRK) of Haynaldia villosa, a diploid wheat relative, has been transferred to wheat variety Yangmai158, which is powdery mildew susceptible [37]. Transgenic wheat plants showed a significant increase in powdery mildew resistance.
A different original approach is represented by engineering novel recombinant PRRs by producing chimeric receptors. Modular assemblies between Arabidopsis EFR and rice Xa21 [47] have shown that it is reliable to engineer PRRs to induce signaling and quantitative immunity against the bacterium Pseudomonas syringae pv. Tomatoe and Agrobacterium tumefaciens in Arabidopsis.
For bacterial pathogens expressing transcription activator-like (TAL) effectors that activate the expression of susceptibility genes in the host, resistance can be engineered introducing deletions in the TAL DNA binding sites on the promoter of those genes [48][49]. Another approach to engineer resistance to these bacterial pathogens is to add TAL effector binding sites to a cell-death-promoting (“executor”) gene that is triggered by TAL effectors present in common pathotypes [50][51].
3.2. Boosting the Immune Signaling
P/DTI and ETI lead to the activation of the membrane-localized ion channels and an increase in the amount of cytoplasmic calcium. Other early response events include the activation of mitogen-activated protein kinases (MAPKs) [52]. Three hormones are principally involved in downstream signaling pathways caused by P/DTI and ETI: SA, jasmonic acid (JA), and ethylene (ET). Even though SA pathway stimulates resistance to biotrophic and hemibiotrophic pathogens, JA and ET pathways are typically induced upon sensing necrotrophic pathogens and chewing insects [53]. JA and SA have important roles in the activation of transcription factors controlling biotic stress responses, the interplay between different defense signaling pathways, and chemical priming to improve plant resistance through systemic acquired resistance (SAR).
Activated defense programs require cellular rearrangements at different levels, including machinery involved in transcription, translation, and protein secretion as well as metabolism prioritization of carbon and nitrogen towards production of defense compounds, such as pathogenesis-related (PR) proteins.
Therefore, the overexpression of specific transcription factors is a potential strategy to engineer resistance, with minimized or no effects on yield. One interesting study concern the rice gene Ideal Plant Architecture 1 (IPA1), known as OsSPL14, in which a naturally occurred allelic variant increased yield and resistance to rice blast (Table 1).
3.3. R Gene Transfer
Adult plant resistance (APR) or “slow rusting” wheat genes represent a class of potential transferable R genes [54]. Different APR genes are known, but only two, Lr34 and Lr67 (Table 1), have been cloned [55][56]. Transgenic wheat lines expressing Lr34 gene displayed enhanced resistance to multiple biotrophic pathogens including the leaf rust pathogen and powdery mildew both at seedling and adult stages [38][57]. The mechanism by which resistance is triggered by Lr34 and Lr67 is poorly understood, although it is likely that it provides the activation of biotic or abiotic stress responses allowing the host to limit pathogen development and growth.
Wheat resistance to Fusarium species has been greatly improved by expressing either a barley uridine diphosphate-dependent glucosyltransferases (UGT), HvUGT13248, involved in mycotoxin detoxification [58], or pyramided inhibitors of cell wall-degrading enzymes secreted by the fungi, such as the bean polygalacturonase inhibiting protein (PvPGIP2) and TAXI-III, a xylanase inhibitor [59].
4. Increasing Disease-Resistance in Cereals by Using Gene Expression or Editing Techniques
4.1. RNA Interference (RNAi)
RNA interference (RNAi) was first discovered in plants as a molecular mechanism involved in the recognition and degradation of non-self-nucleic acids, principally directed against virus-derived sequences. In addition to its defensive role, RNAi is essential for endogenous gene expression regulation [60]. RNAi-based resistance can be engineered against many viruses by expressing “hairpin” structures, double-stranded RNA molecules that contain viral sequences, or simply by overexpressing dysfunctional viral genes [61]. Moreover, a single double-stranded RNA molecule can be processed into a variety of small interfering (si)RNAs and thereby effectively target several virus sequences using a single hairpin construct.
Over the last two decades, RNAi has emerged as a powerful genetic tool for scientific research. In addition to basic studies on the determination of gene function, RNA-silencing technology has been used to develop plants with increased resistance to biotic stresses (Figure 2), (Table 2) [62][63].
In short, RNAi appears to be a promising additional control strategy in the arsenal of plant
breeders against at least some pathogens.
| Molecular Technique | Biotechnological Intervention | Gene | Species | Enhanced Resistance to | References |
|---|---|---|---|---|---|
| RNAi | Viral gene silencing | Wheat streak mosaic virus genes | Wheat | Wheat streak mosaic virus (WSMV) | [64] |
| Wheat dwarf virus genes | Barely | Wheat dwarf virus (WDV) | [65] | ||
| Host-induced gene silencing | FgCYP51A, FgCYP51B and FgCYP51C | Barely | Fusarium graminearum | [66] | |
| FgCh3b | Wheat | Fusarium graminearum | [67] | ||
| PtMAPK1, PtCYC1, PtCNB | Wheat | Puccinia triticina, P. graminis and P. striiformis | [68][69] | ||
| FcGls | Wheat | Fusarium culmorum | [70] | ||
| CRISPR/Cas9 | Silencing of host genes | TaMlo-A1 | Wheat | Blumeria graminis f. sp. tritici | [71] |
| OsSWEET13 | Rice | Xanthomonas oryzae pv. oryzae | [72] | ||
| OsERF922 | Rice | Magnaporthe oryzae | [73] | ||
| TaEDR1 | Wheat | Blumeria graminis f. sp. tritici | [74] | ||
| OsSEC3A | Rice | Magnaporthe oryzae | [75] | ||
| TaLpx-1 | Wheat | Fusarium graminearum | [46] | ||
| TaHRC | Wheat | Fusarium graminearum | [76] |
4.2. CRISPR/Cas9 Mediated Genome Editing
In plant research, NBTs are attracting a lot of attention. NBTs appear to be suitable for many different fields in plant science, such as developmental processes and adaptation/resistance to (a)biotic stresses [77]. NBTs include the most recent and powerful molecular approaches for precise genetic modifications of single or multiple gene targets. They employ site-directed nucleases to introduce double-strand breaks at predetermined sites in DNA.
The rapid increase in scientific publications documenting the use of CRISPR/Cas highlights how this technique has a greater success rate in gene modification compared to the other available nucleases. Actually, the application of CRISPR/Cas technologies to edit plant genomes is proving to be a powerful tool for future enhancement of agronomic traits in crops, qualitative and health parameters, tolerance to abiotic stress [78], and also for the improvement of biotic stress resistance (Table 2) [79].
MLO loci have been targeted by RNA-guided Cas9 endonuclease inbread wheat [80]. It had previously been reported that MLO were susceptibility genes and that homozygous loss-of-function mutants had significantly increased resistance to powdery mildew in barley, Arabidopsis, and tomato [81][82][83]. Bread wheat plants mutated by CRISPR/Cas9 in one (TaMLO-A1) of the three MLO homeoalleles showed improved resistance to Blumeria graminis f. sp. tritici infection. Another example of CRISPR/Cas9-derived resistance against the same disease is the knockout of TaEDR1 [84], conferring resistance to powdery mildew in wheat.
Plants resistant to rice blast disease were generated through CRISPR/Cas9-mediated disruption of OsERF922 and OsSEC3A genes in rice.
Relatively few studies have been published on the application of the CRISPR/Cas systems to counteract crop bacterial diseases. CRISPR/Cas9 editing of OsSWEET13 has been performed in rice to achieve resistance to bacterial blight disease caused by bacterium Xanthomonas oryzae pv. oryzae [72]. X. oryzae produces an effector protein, PthXo2, which induces OsSWEET13 expression in the host and the consequent condition of susceptibility. Zhou et al. [85] obtained a null mutation in OsSWEET13 in order to better explore PthXo2-dependent disease susceptibility, and resultant mutants were resistant to bacterial blight.
Further genome editing strategies for multiplexed recessive resistance using a combination of the major effectors and other R genes will be the next step toward achieving bacterial blight resistance.
This entry is adapted from the peer-reviewed paper 10.3390/plants10061146
References
- Shiferaw, B.; Smale, M.; Braun, H.-J.; Duveiller, E.; Reynolds, M.P.; Muricho, G. Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur. 2013, 5, 291–317.
- Hawkins, N.J.; Bass, C.; Dixon, A.; Neve, P. The evolutionary origins of pesticide resistance. Biol. Rev. 2019, 94, 135–155.
- Pandey, P.; Irulappan, V.; Bagavathiannan, M.V.; Senthil-Kumar, M. Impact of Combined Abiotic and Biotic Stresses on Plant Growth and Avenues for Crop Improvement by Exploiting Physio-morphological Traits. Front. Plant Sci. 2017, 8, 537.
- Gull, A.; Lone, A.A.; Wani, N.U.I. Biotic and Abiotic Stresses in Plants. Abiotic Biot. Stress Plants 2019.
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E.; A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821, .
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nat. Cell Biol. 2019, 576, 149–157.
- Camerlengo, F.; Frittelli, A.; Sparks, C.; Doherty, A.; Martignago, D.; Larré, C.; Lupi, R.; Sestili, F.; Masci, S.; CRISPR-Cas9 Multiplex Editing of the -Amylase/Trypsin Inhibitor Genes to Reduce Allergen Proteins in Durum Wheat.. Front. Sustain. Food Syst. , , 2020, 4, 104, .
- Sun, Y.; Zhang, X.; Wu, C.; He, Y.; Ma, Y.; Hou, H.; Guo, X.; Du, W.; Zhao, Y.; Xia, L.; et al. Engineering Herbicide-Resistant Rice Plants through CRISPR/Cas9-Mediated Homologous Recombination of Acetolactate Synthase. Mol. . Plant 2016, 9, 628–631, .
- León, S.S.; Gil-Humanes, J.; Ozuna, C.V.; Giménez, M.J.; Sousa, C.; Voytas, D.; Barro, F.; Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. . Plant Biotechnol. J. 2017, 16, 902–910, .
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S.; et al. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. . Nat. Biotechnol. 2015, 33, 1162–1164, .
- Kawashima, C.G.; Guimarães, G.A.; Nogueira, S.R.; MacLean, D.; Cook, D.R.; Steuernagel, B.; Baek, J.; Bouyioukos, C.; Melo, B.D.V.A.; Tristão, G.; et al. A pigeonpea gene confers resistance to Asian soybean rust in soybean. Nat. Biotechnol. 2016, 34, 661–665.
- Claus, L.A.N.; Savatin, D.V.; Russinova, E. The crossroads of receptor-mediated signaling and endocytosis in plants. J. Integr. Plant Biol. 2018, 60, 827–840.
- Savatin, D.V.; Gramegna, G.; Modesti, V.; Cervone, F. Wounding in the plant tissue: The defense of a dangerous passage. Front. Plant Sci. 2014, 5, 470.
- Spoel, S.H.; Dong, X. How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 2012, 12, 89–100.
- Dodds, P.N.; Rathjen, J. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548.
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; He, S.Y.; Zhou, J.M.; Xin, X.F. Pat-tern-Recognition Receptors Are Required for NLR-Mediated Plant Immunity. Nature 2020, 592, 105–109.
- Ngou, B.P.M.; Ahn, H.-K.; Ding, P.; Jones, J.D.G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nat. Cell Biol. 2021, 592, 1–6.
- Weyers, J.D.B.; Paterson, N.W. Plant hormones and the control of physiological processes. New Phytol. 2001, 152, 375–407.
- Del Bianco, M.; Giustini, L.; Sabatini, S. Spatiotemporal changes in the role of cytokinin during root development. New Phytol. 2013, 199, 324–338.
- Depuydt, S.; Hardtke, C.S. Hormone Signalling Crosstalk in Plant Growth Regulation. Curr. Biol. 2011, 21, R365–R373.
- Vanstraelen, M.; Benková, E. Hormonal Interactions in the Regulation of Plant Development. Annu. Rev. Cell Dev. Biol. 2012, 28, 463–487.
- Bargmann, B.; Vanneste, S.; Krouk, G.; Nawy, T.; Efroni, I.; Shani, E.; Choe, G.; Friml, J.; Bergmann, D.C.; Estelle, M.; et al. A map of cell type-specific auxin responses. Mol. Syst. Biol. 2013, 9, 688.
- Schoonbeek, H.; Wang, H.; Stefanato, F.L.; Craze, M.; Bowden, S.; Wallington, E.; Zipfel, C.; Ridout, C.J. Arabidopsis EF -Tu receptor enhances bacterial disease resistance in transgenic wheat. New Phytol. 2015, 206, 606–613.
- Schwessinger, B.; Bahar, O.; Thomas, N.; Holton, N.; Nekrasov, V.; Ruan, D.; Canlas, P.E.; Daudi, A.; Petzold, C.; Singan, V.R.; et al. Transgenic Expression of the Dicotyledonous Pattern Recognition Receptor EFR in Rice Leads to Ligand-Dependent Activation of Defense Responses. PLoS Pathog. 2015, 11, e1004809.
- Lu, F.; Wang, H.; Wang, S.; Jiang, W.; Shan, C.; Li, B.; Yang, J.; Zhang, S.; Sun, W. Enhancement of innate immune system in monocot rice by transferring the dicotyledonous elongation factor Tu receptor EFR. J. Integr. Plant Biol. 2014, 57, 641–652.
- Peng, H.; Chen, Z.; Fang, Z.; Zhou, J.; Xia, Z.; Gao, L.; Chen, L.; Li, L.; Li, T.; Zhai, W.; et al. Rice Xa21 primed genes and pathways that are critical for combating bacterial blight infec-tion. Sci Rep. 2015, 5, 12165.
- Chen, T.; Xiao, J.; Xu, J.; Wan, W.; Qin, B.; Cao, A.; Chen, W.; Xing, L.; Du, C.; Gao, X.; et al. Two members of TaRLK family confer powdery mildew resistance in common wheat. BMC Plant Biol. 2016, 16, 27.
- Rajaraman, J.; Douchkov, D.; Hensel, G.; Stefanato, F.L.; Gordon, A.; Ereful, N.; Caldararu, O.F.; Petrescu, A.-J.; Kumlehn, J.; Boyd, L.A.; et al. An LRR/Malectin Receptor-Like Kinase Mediates Resistance to Non-adapted and Adapted Powdery Mildew Fungi in Barley and Wheat. Front. Plant Sci. 2016, 7, 1836.
- Wang, Z.; Cheng, J.; Fan, A.; Zhao, J.; Yu, Z.; Li, Y.; Wang, X. LecRK-V, an L-type lectin receptor kinase in Haynaldia villosa, plays positive role in resistance to wheat powdery mildew. Plant Biotechnol. J. 2018, 16, 50–62.
- Holton, N.; Nekrasov, V.; Ronald, P.C.; Zipfel, C. The Phylogenetically-Related Pattern Recognition Receptors EFR and XA21 Recruit Similar Immune Signaling Components in Monocots and Dicots. PLoS Pathog. 2015, 11, e1004602.
- Thomas, N.C.; Oksenberg, N.; Liu, F.; Caddell, D.; Nalyvayko, A.; Nguyen, Y.; Schwessinger, B.; Ronald, P.C. The rice XA21 ectodomain fused to the Arabidopsis EFR cytoplasmic domain confers resistance to Xanthomonas oryzae pv. oryzae. PeerJ 2018, 6, e4456.
- Boutrot, F.; Zipfel, C. Function, Discovery, and Exploitation of Plant Pattern Recognition Receptors for Broad-Spectrum Disease Resistance. Annu. Rev. Phytopathol. 2017, 55, 257–286.
- Li, T.; Liu, B.; Spalding, M.; Weeks, D.; Yang,, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 2012, 30, 390–392.
- Hummel, A.W.; Doyle, E.L.; Bogdanove, A.J. Addition of transcription activator-like effector binding sites to a pathogen strain-specific rice bacterial blight resistance gene makes it effective against additional strains and against bacterial leaf streak. New Phytol. 2012, 195, 883–893.
- Xu, G.; Yuan, M.; Ai, C.; Liu, L.; Zhuang, E.; Karapetyan, S.; Wang, S.; Dong, X. uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nat. Cell Biol. 2017, 545, 491–494.
- Zhang, Z.; Liu, X.; Wang, X.; Zhou, M.; Zhou, X.; Ye, X.; Wei, X. An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytol. 2012, 196, 1155–1170.
- Wang, J.; Zhou, L.; Shi, H.; Chern, M.; Yu, H.; Yi, H.; He, M.; Yin, J.; Zhu, X.; Li, Y.; et al. A single transcription factor promotes both yield and immunity in rice. Science 2018, 361, 1026–1028.
- Risk, J.M.; Selter, L.L.; Chauhan, H.; Krattinger, S.G.; Kumlehn, J.; Hensel, G.; Viccars, L.A.; Richardson, T.M.; Buesing, G.; Troller, A.; et al. The wheat Lr34 gene provides resistance against multiple fungal pathogens in barley. Plant Biotechnol. J. 2013, 11, 847–854.
- Krattinger, S.G.; Sucher, J.; Selter, L.L.; Chauhan, H.; Zhou, B.; Tang, M.; Upadhyaya, N.M.; Mieulet, D.; Guiderdoni, E.; Weidenbach, D.; et al. The wheat durable, multipathogen resistance geneLr34confers partial blast resistance in rice. Plant Biotechnol. J. 2016, 14, 1261–1268.
- Schnippenkoetter, W.; Lo, C.; Liu, G.; Dibley, K.; Chan, W.L.; White, J.; Milne, R.; Zwart, A.; Kwong, E.; Keller, B.; et al. The wheat Lr34 multipathogen resistance gene confers resistance to anthracnose and rust in sorghum. Plant Biotechnol. J. 2017, 15, 1387–1396.
- Sucher, J.; Boni, R.; Yang, P.; Rogowsky, P.; Büchner, H.; Kastner, C.; Kumlehn, J.; Krattinger, S.G.; Keller, B. The durable wheat disease resistance geneLr34confers common rust and northern corn leaf blight resistance in maize. Plant Biotechnol. J. 2016, 15, 489–496.
- Rinaldo, A.; Gilbert, B.; Boni, R.; Krattinger, S.G.; Singh, D.; Park, R.F.; Lagudah, E.; Ayliffe, M. TheLr34adult plant rust resistance gene provides seedling resistance in durum wheat without senescence. Plant Biotechnol. J. 2017, 15, 894–905.
- Milne, R.J.; Dibley, K.E.; Schnippenkoetter, W.; Mascher, M.; Lui, A.C.; Wang, L.; Lo, C.; Ashton, A.R.; Ryan, P.R.; Lagudah, E.S. The Wheat Lr67 Gene from the Sugar Transport Protein 13 Family Confers Multipathogen Resistance in Barley. Plant Physiol. 2019, 179, 1285–1297.
- Albert, I.; Böhm, H.; Albert, M.; Feiler, C.E.; Imkampe, J.; Wallmeroth, N.; Brancato, C.; Raaymakers, T.M.; Oome, S.; Zhang, H.; et al. An RLP23–SOBIR1–BAK1 complex mediates NLP-triggered immunity. Nat. Plants 2015, 1, 15140.
- Steuernagel, B.; Periyannan, S.K.; Hernández-Pinzón, I.; Witek, K.; Rouse, M.N.; Yu, G.; Hatta, A.; Ayliffe, M.; Bariana, H.; Jones, J.D.G.; et al. Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 2016, 34, 652–655.
- Wang, W.; Pan, Q.; He, F.; Akhunova, A.; Chao, S.; Trick, H.; Akhunov, E. Transgenerational CRISPR-Cas9 Activity Facilitates Multiplex Gene Editing in Allopolyploid Wheat. CRISPR J. 2018, 1, 65–74.
- Holton, N.; Nekrasov, V.; Ronald, P.C.; Zipfel, C.; The Phylogenetically-Related Pattern Recognition Receptors EFR and XA21 Recruit Similar Immune Signaling Components in Monocots and Dicots. . PLoS Pathog. 2015, 11, e1004602, .
- Boutrot, F.; Zipfel, C.; Function, Discovery, and Exploitation of Plant Pattern Recognition Receptors for Broad-Spectrum Disease Resistance. . Annu. Rev. Phytopathol. 2017, 55, 257–286, .
- Jia, H.; Zhang, Y.; Orbovi´c, V.; Xu, J.; White, F.F.; Jones, J.B.; Wang, N.; Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. . Plant Biotechnol. J. 2017, 15, 817–823, .
- Hummel, A.W.; Doyle, E.L.; Bogdanove, A.J.; Addition of transcription activator-like effector binding sites to a pathogen strainspecific rice bacterial blight resistance gene makes it effective against additional strains and against bacterial leaf streak.. New Phytol. 2012, 195, 883–893, .
- Wang, J.; Zhou, L.; Shi, H.; Chern, M.; Yu, H.; Yi, H.; He, M.; Yin, J.; Zhu, X.; Li, Y.; et al. A single transcription factor promotes both yield and immunity in rice.. Science 2018, 361, 1026–1028, .
- Muthamilarasan, M.; Prasad, M. Plant innate immunity: An updated insight into defense mechanism. J. Biosci. 2013, 38, 433–449.
- Vleesschauwer, D.E.; Exu, J.; Hãfte, M. Making sense of hormone-mediated defense networking: From rice to Arabidopsis. Front. Plant Sci. 2014, 5, 611.
- Huerta-Espino, J.; Singh, R.; Crespo-Herrera, L.A.; Villaseñor-Mir, H.E.; Rodriguez-Garcia, M.F.; Dreisigacker, S.; Barcenas-Santana, D.; Lagudah, E. Adult Plant Slow Rusting Genes Confer High Levels of Resistance to Rusts in Bread Wheat Cultivars from Mexico. Front. Plant Sci. 2020, 11, 824.
- Krattinger, S.G.; Lagudah, E.S.; Spielmeyer, W.; Singh, R.P.; Huerta-espino, J.; Mcfadden, H.; Bossolini, E.; Selter, L.L.; Keller, B. Pathogens in Wheat. Science 2009, 323, 1360–1363.
- Ellis, J.G.; Lagudah, E.S.; Spielmeyer, W.; Dodds, P.N. The past, present and future of breeding rust resistant wheat. Front. Plant Sci. 2014, 5, 641.
- Böni, R.H. Functional Characterization of the Wheat Disease Resistance Gene Lr34 in Functional Characterization of the Wheat Disease Resistance Gene Lr34 in Heterologous Barley. Ph.D. Thesis, University of Zurich, Zürich, Switzerland, 2017.
- Mandalà, G.; Tundo, S.; Francesconi, S.; Gevi, F.; Zolla, L.; Ceoloni, C.; D’Ovidio, R.; Deoxynivalenol Detoxification in Transgenic Wheat Confers Resistance to Fusarium Head Blight and Crown Rot Diseases. . Mol. Plant-Microbe Interact. 2019, 32, 583–592, .
- Tundo, S.; Kalunke, R.; Janni, M.; Volpi, C.; Lionetti, V.; Bellincampi, D.; Favaron, F.; D’Ovidio, R.; Pyramiding PvPGIP2 and TAXI-III but Not PvPGIP2 and PMEI Enhances Resistance Against Fusarium graminearum.. Mol. Plant-Microbe Interact. 2016, 29, 629–639, .
- Obbard, D.J.; Gordon, K.H.J.; Buck, A.; Jiggins, F.M. The evolution of RNAi as a defence against viruses and transposable elements. Philos. Trans. R. Soc. B Biol. Sci. 2008, 364, 99–115.
- Rosa, C.; Kuo, Y.-W.; Wuriyanghan, H.; Falk, B.W. RNA Interference Mechanisms and Applications in Plant Pathology. Annu. Rev. Phytopathol. 2018, 56, 581–610.
- Sidahmed, A.M.E.; Wilkie, B. Endogenous Antiviral Mechanisms of RNA Interference: A Comparative Biology Perspective. Adv. Struct. Saf. Stud. 2010, 623, 3–19.
- Gaffar, F.Y.; Koch, A. Catch Me If You Can! RNA Silencing-Based Improvement of Antiviral Plant Immunity. Viruses 2019, 11, 673.
- Fahim, M.; Millar, A.; Wood, C.C.; Larkin, P.J. Resistance to Wheat streak mosaic virus generated by expression of an artificial polycistronic microRNA in wheat. Plant Biotechnol. J. 2011, 10, 150–163.
- Kis, A.; Tholt, G.; Ivanics, M.; Várallyay, É.; Jenes, B.; Havelda, Z. Polycistronic artificial miRNA-mediated resistance toWheat dwarf virusin barley is highly efficient at low temperature. Mol. Plant Pathol. 2015, 17, 427–437.
- Koch, A.; Kumar, N.; Weber, L.; Keller, H.; Imani, J.; Kogel, K.-H. Host-induced gene silencing of cytochrome P450 lanosterol C14 -demethylase-encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. USA 2013, 110, 19324–19329.
- Cheng, W.; Song, X.-S.; Xiao-Li, Q.; Cao, L.-H.; Sun, K.; Qiu, X.-L.; Xu, Y.-B.; Yang, P.; Huang, T.; Zhang, J.-B.; et al. Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 2015, 13, 1335–1345.
- Panwar, V.; McCallum, B.; Bakkeren, G. Endogenous silencing of P uccinia triticina pathogenicity genes through in planta-expressed sequences leads to the suppression of rust diseases on wheat. Plant J. 2013, 73, 521–532.
- Panwar, V.; McCallum, B.; Bakkeren, G. Host-induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the Barley stripe mosaic virus. Plant Mol. Biol. 2013, 81, 595–608.
- Chen, W.; Kastner, C.; Nowara, D.; Oliveira-Garcia, E.; Rutten, T.; Zhao, Y.; Deising, H.B.; Kumlehn, J.; Schweizer, P. Host-induced silencing of Fusarium culmorum genes protects wheat from infection. J Exp Bot. 2016, 67, 4979–4991.
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.-L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951.
- Zhou, J.; Peng, Z.; Long, J.; Sosso, D.; Liu, B.; Eom, J.-S.; Huang, S.; Liu, S.; Cruz, C.V.; Frommer, W.; et al. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 2015, 82, 632–643.
- Wang, F.; Wang, C.; Liu, P.; Lei, C.; Hao, W.; Gao, Y.; Liu, Y.-G.; Zhao, K. Enhanced Rice Blast Resistance by CRISPR/Cas9-Targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PLoS ONE 2016, 11, e0154027.
- Zhang, Y.; Bai, Y.; Wu, G.; Zou, S.; Chen, Y.; Gao, C.; Tang, D. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017, 91, 714–724.
- Ma, J.; Chen, J.; Wang, M.; Ren, Y.; Wang, S.; Lei, C.; Cheng, Z. Sodmergen Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice. J. Exp. Bot. 2018, 69, 1051–1064.
- Su, Z.; Bernardo, A.; Tian, B.; Chen, H.; Wang, S.; Ma, H.; Cai, S.; Liu, D.; Zhang, D.; Li, T.; et al. A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat. Genet. 2019, 51, 1099–1105.
- Nelson, R.; Wiesner-Hanks, T.; Wisser, R.; Balint-Kurti, P. Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet. 2018, 19, 21–33.
- Mohanta, T.K.; Bashir, T.; Hashem, A.; Allah, E.F.A.; Bae, H. Genome Editing Tools in Plants. Genes 2017, 8, 399.
- Arora, L.; Narula, A. Gene Editing and Crop Improvement Using CRISPR-Cas9 System. Front. Plant Sci. 2017, 8, 1932.
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.-L.; Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. . Nat. Biotechnol. 2014, 32, 947–951, .
- Piffanelli, P.; Ramsay, L.; Waugh, R.; Benabdelmouna, A.; D’Hont, A.; Hollricher, K.; Jørgensen, J.H.; Schulze-Lefert, P.; Panstruga, R.; A barley cultivation-associated polymorphism conveys resistance to powdery mildew. . Nat. Cell Biol. 2004, 430, 887–891, .
- Consonni, C.; Humphry, M.E.; Hartmann, H.A.; Livaja, M.; Durner, J.; Westphal, L.; Vogel, J.; Lipka, V.; Kemmerling, B.; Schulze-Lefert, P.; et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. . Nat. Genet. 2006, 38, 716–720, .
- Bai, Y.; Pavan, S.; Zheng, Z.; Zappel, N.F.; Reinstädler, A.; Lotti, C.; De Giovanni, C.; Ricciardi, L.; Lindhout, P.; Visser, R.; et al. Naturally Occurring Broad-Spectrum Powdery Mildew Resistance in a Central American Tomato Accession Is Caused by Loss of Mlo Function. Mol. . Plant-Microbe Interact. 2008, 21, 30–39, .
- Zhang, Y.; Bai, Y.; Wu, G.; Zou, S.; Chen, Y.; Gao, C.; Tang, D.; Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. . Plant J. 2017, 91, 714–724, .
- Junhui Zhou; Zhao Peng; Juying Long; Davide Sosso; Bo Liu; Joon-Seob Eom; Sheng Huang; Sanzhen Liu; Casiana Vera Cruz; Wolf Frommer; et al. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. The Plant Journal 2015, 82, 632-643, 10.1111/tpj.12838.
