Pattern recognition receptors (PRRs) play a crucial role in inducing inflammatory responses; they recognize pathogen-associated molecular patterns, damage-associated molecular patterns, and environmental factors.
- pattern recognition
- pyroptosis
- inflammasome
1. Introduction
Inflammatory responses play a crucial role in the innate immune system to protect the host body from pathogens or other environmental factors. Pattern recognition systems act as inducers of inflammation [1][2]. For activation of the pattern recognition system, pattern recognition receptors (PRRs) recognize pathogen-associated molecular patterns (PAMPs) (e.g., lipopolysaccharide (LPS) and flagellin), damage-associated molecular patterns (DAMPs) (e.g., high mobility group box 1 (HMGB1), adenosine triphosphate (ATP)), or even environmental factors (e.g., particulate silica [3] and aluminum salt [4]) [1][2][5][6]. In the host cell cytosol, nucleotide-binding oligomerization domain (NOD)-leucine-rich repeats (LRR)-containing receptors (NLRs), part of the PRR family, recognize PAMPs, DAMPs, and environmental factors while forming a multiple-protein complex, called the “inflammasome” [1][7][8][9]. The formation and activation of the inflammasomes induce precursor inflammatory caspases (Casps), such as pro-Casp1, that activate Casp1, which then leads to non-apoptotic cell death, called “pyroptosis” [10][11][12][13]. Moreover, Casp1 induces pyroptosis and triggers precursor inflammatory cytokines to activate interleukin (IL)-1β and IL-18 [13][14][15]. These cytokines are secreted into the extracellular space through the pores formed after pyroptosis; furthermore, inflammation protected the host body from pathogens and environmental factors [14][15][16].
2. Inflammasome Activation to Induce Pyroptosis
2.1. Generic Structure of Inflammasome and Mechanisms of Pyroptosis
The inflammasome consists of an NLR (as a sensor protein), an ASC (apoptosis-associated speck-like protein containing a caspase recruitment domain (CARD), also known as pycard or the target of methylation-induced silencing-1 (TMS-1) as an adaptor protein), and pro-Casp1 (an inflammatory caspase) [16][17]. Inflammasome-forming NLRs have an N-terminal effector domain (i.e., pyrin domain (PYD) or CARD) that binds to ASC, a central fish-specific NACHT-associated domain and NACHT domain, and a C-terminal LRR motif that recognizes PAMPs, DAMPs, or environmental factors as ligands [6][9]. Several NLRs recruit the adaptor protein ASC (containing PYD and CARD) and pro-Casp1 to form a multiple-protein complex [16][17][18][19]. After recruitment of ASC and pro-Casp1, pro-Casp1 is self-proteolyzed into its active-form, Casp1 [13]. Casp1 is one of the proteases that cleaves the precursor inflammatory cytokines (i.e., pro-IL-1β and pro-IL-18) into their activated forms IL-1β and IL-18 [13][14][20]. Casp1 also cleaves gasdermin (GSDM) family proteins (i.e., GSDMA, -C, -D, and -E) at the N-terminus of GSDM, which then forms a pore in the cell membrane. Extracellular water then flows into the cell through the GSDM pore; thus, the cell expands. Eventually, the cell membrane is broken, leading to lytic cell death, known as pyroptosis [21][22]; moreover, at this point, cytosolic inflammatory molecules (including IL-1β, IL-18, HMGB1, and ATP) are released into the extracellular space [10][23][24][25]. In mammals, GSDMD induces pyroptosis in a Casp1-dependent manner [26]; however, there is no gsdmd in the fish genome. Current reports demonstrate that GSDME plays a key role in fish pyroptosis, as GSDMD does in mammals [27][28][29]. In zebrafish, CaspA (also known as CaspyA and Caspy) and CaspB (also known as CaspyB and Caspy2) [30], which are orthologs of Casp1, cleave the proinflammatory cytokines and two GSDMEs [27][29].
2.2. The Composition for Inflammasome and Apoptosis-Associated Speck-Like Protein Containing a Caspase Recruitment Domain Formation
2.2.1. Inflammasome Formation
NLRs are components of the inflammasome, and they play a crucial role in recognizing PAMPs, DAMPs, and environmental factors [9]. In mammals, NLRs containing PYD (NLRP1), NLRP3, NLRP12, and CARD (NLRC4) are key sensory molecules in inflammasomes [13][31][32][33][34]. However, in teleosts, only NLRP1 and NLRP3 in zebrafish and NLRP3 in the Japanese flounder have been identified as inflammasome-forming NLRs [27][35][36]. Interestingly, Japanese medaka and zebrafish have many NLRs that belong to NLRC3 and NLRP12 families (67 in Japanese medaka and 43 in zebrafish) in the genome database, although most lack PYD or CARD, which are important in inflammasome formation [37] (Figure 1). In the phylogenetic analysis, Japanese medaka NLRP12 (NLRP12 -3), zebrafish NLRP3, and three zebrafish NLRC3-like (i.e., NLRC3-like 1, NLRC3-4, and NLRC3-7) have PYD, which is divergent from the teleost NLRC3 group (Figure 1). Furthermore, in mammals, NLRC3 consists of only NACHT and LRR. However, mammals also have PYD, DD, FISNA, RING, PRY, and SPRY, in addition to the NACHT and LRR found in the Japanese medaka and NLRC3 and NLRP12 families found in zebrafish, which have branched from the mammalian NLRC3 group.
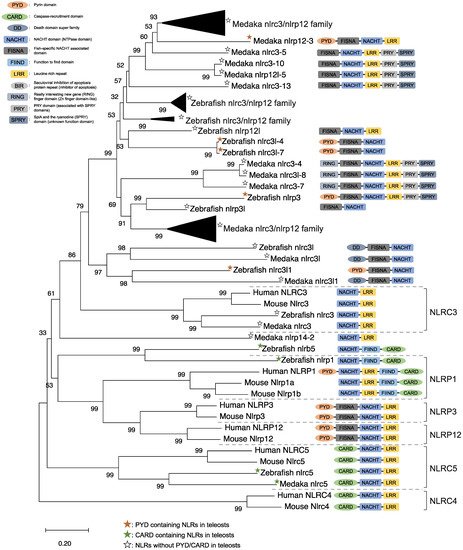
Figure 1. Phylogenetic relationship of the nucleotide-binding oligomerization domain-leucine-rich repeat-containing receptor family in mammals (human and mouse) and fish (Japanese medaka and zebrafish). The tree was constructed using the MEGAX and a neighbor-joining method with 1000 bootstrap replications. The black triangle shapes show the collection of the clusters.
In mice, NLRC3 inhibits TLR signaling by suppressing the signaling adaptor of TRAF6 alongside the transcription factor NF-κB [38]. However, NLRP3, which is clustered with both the zebrafish and Japanese medaka NLRC3 and NLRP12 families (Figure 1), activates the inflammatory response as an inflammasome in zebrafish. Additionally, the Japanese flounder NLRC3 involved extracellular ATP-mediated inflammatory responses [39], while Nile tilapia NLRC3 induced NF-κB activity in mammalian cells [40]. Therefore, both the NLRC3 and NLRP12 families in teleosts, which have branched from mammalian NLRC3 (Figure 1), may undergo a variety of unique evolutionary paths and have different functions from mammalian NLRCs. Furthermore, some NLRC3, NLRP12, and NLRP3 in the Japanese medaka and zebrafish conserve PRY and SPRY domains (also known as B30.2 domain), whereas these domains are not conserved in mammalian NLRs [37] (Figure 1). In zebrafish NLRP3, the B30.2 domain did not influence CaspA or CaspB activities; however, both PYD and NACHT influenced CaspA or CaspB activities [27]. According to the domain structures and phylogenetic analysis, there could be uniquely evolved inflammasome-forming NLRs with similar functions in mammals.
ASC is an adaptor protein of the inflammasome, which is recruited after NLRs recognize ligands (Figure 2). The ASC has two functional domains, PYD and CARD, and has been identified in many fish species, including zebrafish and Japanese medaka [41][42][43][44][45][46][47]. ASC binds to NLRPs via PYD–PYD interactions in mammals [48]. Moreover, in zebrafish and Japanese flounder, NLRP3 and ASC are co-localized in the cytosol, and they are detected as small spot signals under a microscope [27][36]. Furthermore, deletion of PYD in the ASC is not co-localized with NLRP3, and CARD deletion does not influence co-localization in either zebrafish or Japanese flounder [27][36]. Therefore, NLRP3 may bind to ASC through PYD–PYD interactions in teleosts and mammals. NLRP1 has two functional domains in humans: the N-terminal PYD and C-terminal CARD domains, which are important in inflammasome formation (Figure 2). However, zebrafish NLRP1 only has a C-terminal CARD, similar to mice [31][35] (Figure 1). The N-terminal PYD of NLRP1 has autolytic activity in function to find domain (FIIND), and it inhibits the formation of the NLRP1 inflammasome in humans [31][32]. As a result, the important inflammasome-forming domain in humans, like in mice, is CARD. In mammals, NLRP1 can recruit ASC pro-Casp1 via CARD–CARD interactions [31] (Figure 2). In contrast, zebrafish NLRP1 recruits only ASC via the CARD–CARD interaction; subsequently, the ASC recruits pro-CaspA or pro-CaspB [35] (Figure 2). The difference in the NLRP1 recruitment molecules between mammals and zebrafish is thought to be related to the structure of Casp1. The differences in the structure of the aforementioned Casp1 proteins are described in the following section. In zebrafish, NLRP1 is co-localized with ASC during spot formation in the cytosol. However, the PYD-deleted ASC is not entirely co-localized with NLRP3 and oligomerizes during filament formation. In contrast, the CARD-deleted ASC does not show any co-localization with NLRP1 in zebrafish [35]. Consequently, zebrafish NLRP1 may bind to ASC via a CARD–CARD interaction (Figure 2). In a recent study, there were three replicated ASCs in Japanese medaka, all of which had PYD and CARD [42]. It is unclear whether these ASCs also bind to NLRP family members, which needs to be investigated in the future.
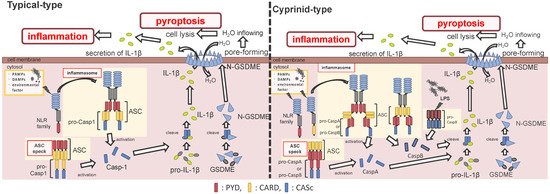
Figure 2. Differences between the cyprinids and other fish species in their inflammasome activation and pyroptosis pathways. In most fish, the nucleotide-binding oligomerization domain-leucine-rich repeat-containing receptor (NLR) family recognizes the ligands, and bind to apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) and pro-Caspase1 (Casp1) via pyrin domain (PYD)-PYD and caspase recruitment domain (CARD)-CARD interactions, thus forming the inflammasome. However, in cyprinids, ligands recognize the NLR family and construct two types of inflammasomes: in the first, the NLR family binds to ASC and pro-CaspA/B via CARD–CARD and PYD–PYD interactions, respectively. The NLR family interacts with ASC via the PYD–PYD interaction, and the ASC oligomerization occurs via CARD–CARD (called ASC core). Then, the pro-CaspA/B binds to the ASC core via the PYD–PYD interaction. After activation, the Casp1/A/B undergoes self-proteolysis, and the Casp1 matures interleukin-1β, inducing pyroptosis through gasdermin-E cleavage in all fish, including cyprinids. In addition, the cyprinid’s CaspB works as a non-canonical inflammasome activator, similar to mammalian Casp4/5/11. The pro-CaspB directly recognizes liposaccharides and thus activates by itself.
After the interaction between NLRs and ASC, ASC then recruits pro-Casp1, and together, they form the inflammasome (Figure 2). In all vertebrates, excluding cyprinid fish, pro-Casp1 has two functional domains, CARD and caspase consensus (CASc) (Table 1). Pro-Casp1 is self-proteolyzed into the active-form Casp1 after it binds with ASC, mutually catalyzing other pro-Casp1s [49][50]. Japanese flounder pro-Casp1 was also activated by NLRP3 [36]. In contrast, cyprinids pro-CaspA and pro-CaspyB possess PYD and CASc domains instead of CARD; moreover, zebrafish pro-CaspA and B are co-localized with NLRP1, NLRP3, and ASC in the spot formation [27][35]. Therefore, in teleosts, pro-Casp1 is activated via NLRP1 or NLRP3 inflammasome formation.
Table 1. Structures of typical and cyprinid-types pro-Caspase 1 in vertebrates.
| Species | Formation Type | Gene Name | Ensembl Gene ID | Domain Structure | |
|---|---|---|---|---|---|
| Mammal | Human (Homo sapiens) |
Typical-type | CASP1 | ENSG00000137752 | 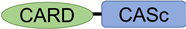 |
| Mouse (Mus musculus) |
Typical-type | Casp1 | ENSMUSG00000025888 |  |
|
| Bird | Chicken (Gallus gallus) |
Typical-type | CASP1 | ENSGALG00000001049 |  |
| Reptile | Common wall lizard (Podarcis muralis) |
Typical-type | CASP1 | ENSPMRG00000020733 |  |
| Amphibian | Tropical clawed frog (Xenopus tropicalis) |
Typical-type | casp1 | ENSXETG00000007792 |  |
| Fish | Japanese medaka (Oryzias latipes) |
Typical-type | casp1 | ENSORLG00000006320 |  |
| Japanese pufferfish (Takifugu rubripes) |
Typical-type | casp1 | ENSTRUG00000007971 |  |
|
| Gilthead seabream (Sparus aurata) |
Typical-type | casp1 | ENSSAUG00010008488 |  |
|
| Turbot (Scophtalmus maximus) |
Typical-type | casp1 | ENSSMAG00000013017 |  |
|
| Zebrafish (Danio rerio) |
Cyprinid-type | caspa | ENSDARG00000008165 |  |
|
| caspb | ENSDARG00000052039 |  |
|||
| caspbl | ENSDARG00000094433 |  |
|||
| Common carp (Cyprinus carpio) |
Cyprinid-type | caspa | ENSCCRG00000035668 |  |
|
| caspb | ENSCCRG00000040063 |  |
To determine the similarity between ASC-binding domains in the vertebrate Casp1 families, the CARD domains of typical Casp1s were compared to the PYD domain in the cyprinid-types. In the alignment and WebLogo analyses, 41 amino acid residues (Met1, Ala2, Asp3, Lys7, Leu9, Arg13, Phe16, Val20, Ile25, Leu28, Leu29, Asp30, Leu32, Leu33, Glu34, Val37, Leu38, Asn39, Glu42, Glu44, Glu50, Asn51, Asp56, Ala58, Arg59, Leu61, Ile62, Asp63, Val65, Lys68, Gly69, Ala72, Ile77, Asp84, Leu87, Leu91, Gly92, Leu93, Thr95, His96, and Ile97) in the general teleost CARD well conserved to those of tetrapods; however, only 8 amino acid residues (Ile25, Leu28, Val37, Asp56, Gly70, Thr93, His94, and Ile95) in the cyprinid PYDs are conserved within the tetrapod CARDs. Furthermore, the 30th Asp (D) residue, related to the interaction of the CARD part of the ASC [49], is conserved in both tetrapod and general teleost CARDs, whereas this conservation does not exist in the cyprinid PYD. Consequently, the ASC-binding domain in the cyprinids showed a unique structure compared to those of the other vertebrates.
2.2.2. Apoptosis-Associated Speck-Like Protein Containing a Caspase Recruitment Domain Formation
This entry is adapted from the peer-reviewed paper 10.3390/ijms22094389
References
- Kawai, T.; Akira, S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009, 21, 317–337.
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820.
- Franchi, L.; Eigenbrod, T.; Núñez, G. Cutting Edge: TNF-α mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J. Immunol. 2009, 183, 792–796.
- Franchi, L.; Núñez, G. The Nlrp3 inflammasome is critical for aluminium hydroxide-mediated IL-1beta secretion but dispensable for adjuvant activity. Eur. J. Immunol. 2008, 38, 2085–2089.
- Brubaker, S.W.; Bonham, K.S.; Zanoni, I.; Kagan, J.C. Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 2015, 33, 257–290.
- Sahoo, B.R. Structure of fish toll-like receptors (TLR) and NOD-like receptors (NLR). Int. J. Biol. Macromol. 2020, 161, 1602–1617.
- Becker, C.E.; O’Neill, L.A.J. Inflammasomes in inflammatory disorders: The role of TLRs and their interactions with NLRs. Semin. Immunopathol. 2007, 29, 239–248.
- Davis, B.K.; Wen, H.; Ting, J.P.-Y. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 2011, 29, 707–835.
- Netea, M.G.; Van de Veerdonk, F.L.; Kullberg, B.J.; Van der Meer, J.W.M.; Joosten, L.A.B. The role of NLRs and TLRs in the activation of the inflammasome. Expert Opin. Biol. Ther. 2008, 8, 1867–1872.
- Bergsbaken, T.; Fink, S.L.; Cookson, B.T. Pyroptosis: Host cell death and inflammation. Nat. Rev. Microbiol. 2009, 7, 99–109.
- Vande Walle, L.; Lamkanfi, M. Pyroptosis. Curr. Biol. 2016, 26, R568–R572.
- Fink, S.L.; Cookson, B.T. Apoptosis, pyroptosis, and necrosis: Mechanistic description of dead and dying eukaryotic cells. Infect. Immun. 2005, 73, 1907–1916.
- Franchi, L.; Eigenbrod, T.; Muñoz-Planillo, R.; Nuñez, G. The inflammasome: A caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat. Immunol. 2009, 10, 241–247.
- Sahoo, M.; Ceballos-Olvera, I.; Del Barrio, L.; Re, F. Role of the inflammasome, IL-1β, and IL-18 in bacterial infections. Sci. World J. 2011, 11, 2037–2050.
- Netea, M.G.; Nold-Petry, C.A.; Nold, M.F.; Joosten, L.A.B.; Opitz, B.; Van Der Meer, J.H.M.; Van De Veerdonk, F.L.; Ferwerda, G.; Heinhuis, B.; Devesa, I.; et al. Differential requirement for the activation of the inflammasome for processing and release of IL-1β in monocytes and macrophages. Blood 2009, 113, 2324–2335.
- Mariathasan, S.; Monack, D.M. Inflammasome adaptors and sensors: Intracellular regulators of infection and inflammation. Nat. Rev. Immunol. 2007, 7, 31–40.
- Lu, A.; Wu, H. Structural mechanisms of inflammasome assembly. FEBS J. 2015, 282, 435–444.
- Guo, H.; Callaway, J.B.; Ting, J.P. Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat. Med. 2015, 21, 677–687.
- Ta, A.; Vanaja, S.K. Inflammasome activation and evasion by bacterial pathogens. Curr. Opin. Immunol. 2021, 68, 125–133.
- Miller, L.S.; Pietras, E.M.; Uricchio, L.H.; Hirano, K.; Rao, S.; Lin, H.; O’Connell, R.M.; Iwakura, Y.; Cheung, A.L.; Cheng, G.; et al. Inflammasome-mediated production of IL-1beta is required for neutrophil recruitment against Staphylococcus aureus in vivo. J. Immunol. 2007, 179, 6933–6942.
- Varela, M.; Romero, A.; Dios, S.; van der Vaart, M.; Figueras, A.; Meijer, A.H.; Novoa, B. Cellular visualization of macrophage pyroptosis and interleukin-1β release in a viral hemorrhagic infection in zebrafish larvae. J. Virol. 2014, 88, 12026–12040.
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665.
- Lopez-Castejon, G.; Brough, D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 2011, 22, 189–195.
- Ceballos-Olvera, I.; Sahoo, M.; Miller, M.A.; del Barrio, L.; Re, F. Inflammasome-dependent pyroptosis and IL-18 protect against Burkholderia pseudomallei lung infection while IL-1β is deleterious. PLoS Pathog. 2011, 7.
- Willingham, S.B.; Allen, I.C.; Bergstralh, D.T.; Brickey, W.J.; Huang, M.T.H.; Taxman, D.J.; Duncan, J.A.; Ting, J.P.Y. NLRP3 (NALP3, cryopyrin) facilitates in vivo caspase-1 activation, necrosis, and HMGB1 release via inflammasome-dependent and -independent pathways. J. Immunol. 2009, 183, 2008–2015.
- Vince, J.E.; Silke, J. The intersection of cell death and inflammasome activation. Cell. Mol. Life Sci. 2016, 73, 2349–2367.
- Li, J.Y.; Wang, Y.Y.; Shao, T.; Fan, D.D.; Lin, A.F.; Xiang, L.X.; Shao, J.Z. The zebrafish NLRP3 inflammasome has functional roles in ASC-dependent interleukin-1β maturation and gasdermin E-mediated pyroptosis. J. Biol. Chem. 2020, 295, 1120–1141.
- Jiang, S.; Gu, H.; Zhao, Y.; Sun, L. Teleost gasdermin E is cleaved by caspase 1, 3, and 7 and induces pyroptosis. J. Immunol. 2019, 203, 1369–1382.
- Wang, Z.; Gu, Z.; Hou, Q.; Chen, W.; Mu, D.; Zhang, Y.; Liu, Q.; Liu, Z.; Yang, D. Zebrafish GSDMEb cleavage-gated pyroptosis drives septic acute kidney injury in vivo. J. Immunol. 2020, 204, 1929–1942.
- Masumoto, J.; Zhou, W.; Chen, F.F.; Su, F.; Kuwada, J.Y.; Hidaka, E.; Katsuyama, T.; Sagara, J.; Taniguchi, S.; Ngo-Hazelett, P.; et al. Caspy, a zebrafish caspase, activated by ASC oligomerization is required for pharyngeal arch development. J. Biol. Chem. 2003, 278, 4268–4276.
- Chavarría-Smith, J.; Vance, R.E. The NLRP1 inflammasomes. Immunol. Rev. 2015, 265, 22–34.
- Schroder, K.; Tschopp, J. The Inflammasomes. Cell 2010, 140, 821–832.
- Cassel, S.L.; Joly, S.; Sutterwala, F.S. The NLRP3 inflammasome: A sensor of immune danger signals. Semin. Immunol. 2009, 21, 194–198.
- Tuladhar, S.; Kanneganti, T.D. NLRP12 in innate immunity and inflammation. Mol. Asp. Med. 2020, 76, 100887.
- Li, J.; Gao, K.; Shao, T.; Fan, D.; Hu, C.; Sun, C.; Dong, W.; Lin, A.; Xiang, L.; Shao, J. Characterization of an NLRP1 inflammasome from zebrafish reveals a unique sequential activation mechanism underlying inflammatory caspases in ancient vertebrates. J. Immunol. 2018, 201, 1946–1966.
- Chen, H.; Ding, S.; Tan, J.; Yang, D.; Zhang, Y.; Liu, Q. Characterization of the Japanese flounder NLRP3 inflammasome in restricting Edwardsiella piscicida colonization in vivo. Fish Shellfish Immunol. 2020, 103, 169–180.
- Chang, M.X.; Xiong, F.; Wu, X.M.; Hu, Y.W. The expanding and function of NLRC3 or NLRC3-like in teleost fish: Recent advances and novel insights. Dev. Comp. Immunol. 2021, 114, 103859.
- Schneider, M.; Zimmermann, A.G.; Roberts, R.A.; Zhang, L.; Swanson, K.V.; Wen, H.; Davis, B.K.; Allen, I.C.; Holl, E.K.; Ye, Z.; et al. The innate immune sensor NLRC3 attenuates toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-κB. Nat. Immunol. 2012, 13, 823–831.
- Li, S.; Chen, X.; Hao, G.; Geng, X.; Zhan, W.; Sun, J. Identification and characterization of a novel NOD-like receptor family CARD domain containing 3 gene in response to extracellular ATP stimulation and its role in regulating LPS-induced innate immune response in Japanese flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2016, 50, 79–90.
- Gao, F.-Y.; Pang, J.-C.; Lu, M.-X.; Yang, X.-L.; Zhu, H.-P.; Ke, X.-L.; Liu, Z.-G.; Cao, J.-M.; Wang, M. Molecular characterization, expression and functional analysis of NOD1, NOD2 and NLRC3 in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2018, 73, 207–219.
- Li, Y.; Huang, Y.; Cao, X.; Yin, X.; Jin, X.; Liu, S.; Jiang, J.; Jiang, W.; Xiao, T.S.; Zhou, R.; et al. Functional and structural characterization of zebrafish ASC. FEBS J. 2018, 285, 2691–2707.
- Morimoto, N.; Okamura, Y.; Kono, T.; Sakai, M.; Hikima, J. Characterization and expression analysis of tandemly-replicated asc genes in the Japanese medaka, Oryzias latipes. Dev. Comp. Immunol. 2020, 115, 103894.
- Xie, J.; Belosevic, M. Functional characterization of apoptosis-associated speck-like protein (ASC) of the goldfish (Carassius auratus L.). Dev. Comp. Immunol. 2016, 65, 201–210.
- Zhang, X.; Liu, Z.; Li, C.; Zhang, Y.; Wang, L.; Wei, J.; Qin, Q. Characterization of orange-spotted grouper (Epinephelus coioides) ASC and caspase-1 involved in extracellular ATP-mediated immune signaling in fish. Fish Shellfish Immunol. 2020, 97, 58–71.
- Sun, Y.; Wang, J.; Lao, H.; Yin, Z.; He, W.; Weng, S.; Yu, X.; Chan, S.M.; He, J. Molecular cloning and expression analysis of the ASC gene from mandarin fish and its regulation of NF-κB activation. Dev. Comp. Immunol. 2008, 32, 391–399.
- Li, S.; Chen, X.; Peng, W.; Hao, G.; Geng, X.; Zhan, W.; Sun, J. Cloning and characterization of apoptosis-associated speck-like protein containing a CARD domain (ASC) gene from Japanese flounder Paralichthys olivaceus. Fish Shellfish Immunol. 2016, 54, 294–301.
- Wang, W.; Tan, J.; Wang, Z.; Zhang, Y.; Liu, Q.; Yang, D. Characterization of the inflammasome component SmASC in turbot (Scophthalmus maximus). Fish Shellfish Immunol. 2020, 100, 324–333.
- Moriya, M.; Taniguchi, S.; Wu, P.; Liepinsh, E.; Otting, G.; Sagara, J. Role of charged and hydrophobic residues in the oligomerization of the pyrin domain of ASC. Biochemistry 2005, 44, 575–583.
- Kersse, K.; Lamkanfi, M.; Bertrand, M.J.M.; Berghe, T.V.; Vandenabeele, P. Interaction patches of procaspase-1 caspase recruitment domains (CARDs) are differently involved in procaspase-1 activation and receptor-interacting protein 2 (RIP2)-dependent nuclear factor κB signaling. J. Biol. Chem. 2011, 286, 35874–35882.
- Srinivasula, S.M.; Poyet, J.L.; Razmara, M.; Datta, P.; Zhang, Z.; Alnemri, E.S. The pyrin-CARD protein ASC is an activating adaptor for caspase-1. J. Biol. Chem. 2002, 277, 21119–21122.
- Hoss, F.; Rodriguez-Alcazar, J.F.; Latz, E. Assembly and regulation of ASC specks. Cell. Mol. Life Sci. 2017, 74, 1211–1229.
- Hara, H.; Tsuchiya, K.; Kawamura, I.; Fang, R.; Hernandez-Cuellar, E.; Shen, Y.; Mizuguchi, J.; Schweighoffer, E.; Tybulewicz, V.; Mitsuyama, M. Phosphorylation of the adaptor ASC acts as a molecular switch that controls the formation of speck-like aggregates and inflammasome activity. Nat. Immunol. 2013, 14, 1247–1255.
- De Alba, E. Structure, interactions and self-assembly of ASC-dependent inflammasomes. Arch. Biochem. Biophys. 2019, 670, 15–31.
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420.
- Kuri, P.; Schieber, N.L.; Thumberger, T.; Wittbrodt, J.; Schwab, Y.; Leptin, M. Dynamics of ASC speck formation during skin inflammatory responses in vivo. bioRxiv 2017, 216.
