Membrane proteins (MPs) are essential for cellular functions. Understanding the functions of MPs is crucial as they constitute an important class of drug targets. However, MPs are a challenging class of biomolecules to analyze because they cannot be studied outside their native environment. Their structure, function and activity are highly dependent on the local lipid environment, and these properties are compromised when the protein does not reside in the cell membrane. Mammalian cell membranes are complex and composed of different lipid species. Model membranes have been developed to provide an adequate environment to envisage MP reconstitution. Among them, tethered-Bilayer Lipid Membranes (tBLMs) appear as the best model because they allow the lipid bilayer to be decoupled from the support. Thus, they provide a sufficient aqueous space to envisage the proper accommodation of large extra-membranous domains of MPs, extending outside. Additionally, as the bilayer remains attached to tethers covalently fixed to the solid support, they can be investigated by a wide variety of surface-sensitive analytical techniques.
- biomimetic membranes
- tethered-Bilayer Lipid Membranes
- membrane proteins
1. Introduction
2. Design of Tethered-Bilayer Lipid Membranes
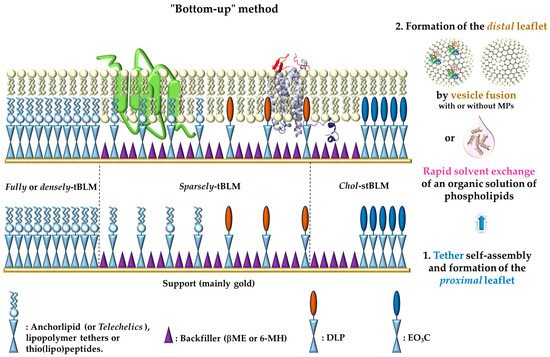
 : represents either anchorlipid (i.e., Telechelics) mainly 2,3-di-O-phytanyl-sn-glycerol-1-tetraethylene glycol-d,l-α-lipoic acid ester lipid (DPhyTL), lipopolymer tethers or thio(lipo)peptides;
: represents either anchorlipid (i.e., Telechelics) mainly 2,3-di-O-phytanyl-sn-glycerol-1-tetraethylene glycol-d,l-α-lipoic acid ester lipid (DPhyTL), lipopolymer tethers or thio(lipo)peptides; 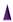 : Backfillers, mainly β-mercaptoethaonl (βME) or 6-mercaptohexanol (6-MH);
: Backfillers, mainly β-mercaptoethaonl (βME) or 6-mercaptohexanol (6-MH); 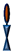 : benzyl-disulfide (tetra-ethyleneglycol)n=2 C20-phytanyl tether (or DLP);
: benzyl-disulfide (tetra-ethyleneglycol)n=2 C20-phytanyl tether (or DLP); 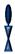 : ethyleneoxy-linked cholesterol (or EO3C).
: ethyleneoxy-linked cholesterol (or EO3C).3. Characterization of Tethered-Bilayer Lipid Membranes
| Techniques | Bilayer Characterization | Surfaces |
|---|---|---|
| Surface plasmon resonance (SPR) imaging | Optical thickness of the bilayer, highly sensitive real-time monitoring of interactions without labeling of the analytes or the ligand, real-time monitoring of bilayer formation | Gold, silver, aluminum |
| Quartz crystal microbalance with dissipation (QCM-D) | Interfacial wet mass determination and viscoelasticity (dissipation sensitive to viscoelastic properties of the adsorbed material), (acoustic) film thickness, real-time monitoring of bilayer formation | Gold, SiO2, mica, metal oxides |
| Imaging ellipsometry (IE) | Indirect quantitative characterization of structural and functional properties of bilayers such as thickness and dry adsorbed mass (i.e., lipids in the adsorbed layer), anisotropy (lateral uniformity and phase separation), molecular area, and receptor-protein interaction affinities. Real-time large area imaging with high sensitivity | Oxide (silicon) substrates |
| Fluorescence recovery after photobleaching (FRAP) | Dynamics, fluidity, and mobility characterisation of lipids and proteins (peripheral or integral), intergrity of artificial membranes | Optically transparent substrates: glass, silica, silcon, gold |
| Electrochemical impedance spectroscopy (EIS) | Electrical properties (resistance and capacitance) of lipid bilayer membranes, formation process in real-time, stability of the membrane, characterization of incorporated ion channels | Gold, silicon |
| Atomic force microscopy (AFM) | In-plane structure and morphology: surface roughness determination, investigation of bilayer surface at the nanoscale range in real-time and in aqueous environment, direct measure of physical properties at high spatial resolution, phase separation (domain formation) and quantification of bilayer thickness | Atomically flat surfaces: mica, silicon, quartz, flat gold |
| (AFM) single-molecule Force Spectroscopy (FS) | Membrane stiffness and mechanical stability on the nanometer length scale, in-depth insight of the orientation of reconstituted transmembrane proteins | Mica, silicon, quartz, flat gold |
| Neutron Reflectometry (NR) | Non-damaging technique giving high structural information on lipid bilayer and internal distribution of components (lipid or protein) within the bilayer (thickness of stratified layers normal to the interface), roughness and interaction with inserted proteins (easy differentiation of lipid and polypeptide components across the membrane structure after interaction) | Gold, silicon |
| X-ray photoelectron spectroscopy (XPS) | Provides quantitaive analysis of elemental composition of a surface and its chemical state | Quartz |
| Grazing incidence small angle neutron or X-ray scattering (GISANS and GISAXS) | Non-destructive method for the structural investigation of biomembranes and mixed lipids systems with different topologies | Performed in quartz glass |
This entry is adapted from the peer-reviewed paper 10.3390/app11114876
References
- Nicolson, G.L. The Fluid—Mosaic Model of Membrane Structure: Still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim. Biophys. Acta Biomembr. 2014, 1838, 1451–1466.
- Liu, J.; Rost, B. Comparing function and structure between entire proteomes. Protein Sci. 2001, 10, 1970–1979.
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124.
- Overington, J.P.; Al-Lazikani, B.; Hopkins, A.L. How many drug targets are there? Nat. Rev. Drug Discov. 2006, 5, 993–996.
- Yin, H.; Flynn, A.D. Drugging Membrane Protein Interactions. Annu. Rev. Biomed. Eng. 2016, 18, 51–76.
- Simons, K.; Ikonen, E. Functional rafts in cell membranes. Nature 1997, 387, 569–572.
- Lingwood, D.; Kaiser, H.-J.; Levental, I.; Simons, K. Lipid rafts as functional heterogeneity in cell membranes. Biochem. Soc. Trans. 2009, 37, 955–960.
- Lingwood, D.; Simons, K. Lipid rafts as a membrane-organizing principle. Science 2010, 327, 46–50.
- Dart, C. SYMPOSIUM REVIEW: Lipid microdomains and the regulation of ion channel function. J. Physiol. 2010, 588, 3169–3178.
- Simons, K.; Sampaio, J.L. Membrane organization and lipid rafts. Cold Spring Harbor Perspect. Biol. 2011, 3, a004697.
- Escribá, P.V. Membrane-lipid therapy: A new approach in molecular medicine. Trends Mol. Med. 2006, 12, 34–43.
- Escribá, P.V.; González-Ros, J.M.; Goñi, F.M.; Kinnunen, P.K.J.; Vigh, L.; Sánchez-Magraner, L.; Fernández, A.M.; Busquets, X.; Horváth, I.; Barceló-Coblijn, G. Membranes: A meeting point for lipids, proteins and therapies. J. Cell. Mol. Med. 2008, 12, 829–875.
- Escribá, P.V.; Busquets, X.; Inokuchi, J.-i.; Balogh, G.; Török, Z.; Horváth, I.; Harwood, J.L.; Vígh, L. Membrane lipid therapy: Modulation of the cell membrane composition and structure as a molecular base for drug discovery and new disease treatment. Prog. Lipid Res. 2015, 59, 38–53.
- Penkauskas, T.; Preta, G. Biological applications of tethered bilayer lipid membranes. Biochimie 2019, 157, 131–141.
- Van den Brink-van der Laan, E.; Antoinette Killian, J.; de Kruijff, B. Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim. Biophys. Acta Biomembr. 2004, 1666, 275–288.
- Dlouhý, O.; Kurasová, I.; Karlický, V.; Javornik, U.; Šket, P.; Petrova, N.Z.; Krumova, S.B.; Plavec, J.; Ughy, B.; Špunda, V.; et al. Modulation of non-bilayer lipid phases and the structure and functions of thylakoid membranes: Effects on the water-soluble enzyme violaxanthin de-epoxidase. Sci. Rep. 2020, 10, 11959.
- Koldsø, H.; Sansom, M.S.P. Local Lipid Reorganization by a Transmembrane Protein Domain. J. Phys. Chem. Lett. 2012, 3, 3498–3502.
- Tamm, L.K.; McConnell, H.M. Supported phospholipid bilayers. Biophys. J. 1985, 47, 105–113.
- Brian, A.A.; McConnell, H.M. Allogeneic stimulation of cytotoxic T cells by supported planar membranes. Proc. Natl. Acad. Sci. USA 1984, 81, 6159–6163.
- Wagner, M.L.; Tamm, L.K. Tethered Polymer-Supported Planar Lipid Bilayers for Reconstitution of Integral Membrane Proteins: Silane-Polyethyleneglycol-Lipid as a Cushion and Covalent Linker. Biophys. J. 2000, 79, 1400–1414.
- Sackmann, E. Supported Membranes: Scientific and Practical Applications. Science 1996, 271, 43–48.
- Sackmann, E.; Tanaka, M. Supported membranes on soft polymer cushions: Fabrication, characterization and applications. Trends Biotechnol. 2000, 18, 58–64.
- Silin, V.I.; Wieder, H.; Woodward, J.T.; Valincius, G.; Offenhausser, A.; Plant, A.L. The Role of Surface Free Energy on the Formation of Hybrid Bilayer Membranes. J. Am. Chem. Soc. 2002, 124, 14676–14683.
- Terrettaz, S.; Mayer, M.; Vogel, H. Highly Electrically Insulating Tethered Lipid Bilayers for Probing the Function of Ion Channel Proteins. Langmuir 2003, 19, 5567–5569.
- Cullison, J.K.; Hawkridge, F.M.; Nakashima, N.; Yoshikawa, S. A Study of Cytochrome c Oxidase in Lipid Bilayer Membranes on Electrode Surfaces. Langmuir 1994, 10, 877–882.
- Ogier, S.D.; Bushby, R.J.; Cheng, Y.; Evans, S.D.; Evans, S.W.; Jenkins, A.T.A.; Knowles, P.F.; Miles, R.E. Suspended Planar Phospholipid Bilayers on Micromachined Supports. Langmuir 2000, 16, 5696–5701.
- Römer, W.; Steinem, C. Impedance Analysis and Single-Channel Recordings on Nano-Black Lipid Membranes Based on Porous Alumina. Biophys. J. 2004, 86, 955–965.
- Römer, W.; Lam, Y.H.; Fischer, D.; Watts, A.; Fischer, W.B.; Göring, P.; Wehrspohn, R.B.; Gösele, U.; Steinem, C. Channel Activity of a Viral Transmembrane Peptide in Micro-BLMs: Vpu1-32 from HIV-1. J. Am. Chem. Soc. 2004, 126, 16267–16274.
- Guidelli, R.; Aloisi, G.; Becucci, L.; Dolfi, A.; Rosa Moncelli, M.; Tadini Buoninsegni, F. Bioelectrochemistry at metal/water interfaces. J. Electroanal. Chem. 2001, 504, 1–28.
- Naumann, R.; Schiller, S.M.; Giess, F.; Grohe, B.; Hartman, K.B.; Kärcher, I.; Köper, I.; Lübben, J.; Vasilev, K.; Knoll, W. Tethered Lipid Bilayers on Ultraflat Gold Surfaces. Langmuir 2003, 19, 5435–5443.
- Lang, H.; Duschl, C.; Vogel, H. A new class of thiolipids for the attachment of lipid bilayers on gold surfaces. Langmuir 1994, 10, 197–210.
- Schiller, S.M.; Naumann, R.; Lovejoy, K.; Kunz, H.; Knoll, W. Archaea Analogue Thiolipids for Tethered Bilayer Lipid Membranes on Ultrasmooth Gold Surfaces. Angew. Chem. Int. Ed. 2003, 42, 208–211.
- Förtig, A.; Jordan, R.; Graf, K.; Schiavon, G.; Purrucker, O.; Tanaka, M. Solid-supported biomimetic membranes with tailored lipopolymer tethers. Macromol. Symp. 2004, 210, 329–338.
- Knoll, W.; Frank, C.W.; Heibel, C.; Naumann, R.; Offenhäusser, A.; Rühe, J.; Schmidt, E.K.; Shen, W.W.; Sinner, A. Functional tethered lipid bilayers. Rev. Mol. Biotechnol. 2000, 74, 137–158.
- Rossi, C.; Homand, J.; Bauche, C.; Hamdi, H.; Ladant, D.; Chopineau, J. Differential Mechanisms for Calcium-Dependent Protein/Membrane Association as Evidenced from SPR-Binding Studies on Supported Biomimetic Membranes†. Biochemistry 2003, 42, 15273–15283.
- Deniaud, A.; Rossi, C.; Berquand, A.; Homand, J.; Campagna, S.; Knoll, W.; Brenner, C.; Chopineau, J. Voltage-Dependent Anion Channel Transports Calcium Ions through Biomimetic Membranes. Langmuir 2007, 23, 3898–3905.
- Yildiz, A.A.; Yildiz, U.H.; Liedberg, B.; Sinner, E.K. Biomimetic membrane platform: Fabrication, characterization and applications. Colloids Surf. B 2013, 103, 510–516.
- Zieleniecki, J.L.; Nagarajan, Y.; Waters, S.; Rongala, J.; Thompson, V.; Hrmova, M.; Köper, I. Cell-Free Synthesis of a Functional Membrane Transporter into a Tethered Bilayer Lipid Membrane. Langmuir 2016, 32, 2445–2449.
- Coutable, A.; Thibault, C.; Chalmeau, J.; François, J.M.; Vieu, C.; Noireaux, V.; Trévisiol, E. Preparation of Tethered-Lipid Bilayers on Gold Surfaces for the Incorporation of Integral Membrane Proteins Synthesized by Cell-Free Expression. Langmuir 2014, 30, 3132–3141.
- Clifton, L.A.; Campbell, R.A.; Sebastiani, F.; Campos-Terán, J.; Gonzalez-Martinez, J.F.; Björklund, S.; Sotres, J.; Cárdenas, M. Design and use of model membranes to study biomolecular interactions using complementary surface-sensitive techniques. Adv. Colloid Interface Sci. 2020, 277, 102118.
- A Biosensor That Uses Ion-Channel Switches. Available online: (accessed on 26 May 2021).
- Jackman, J.; Knoll, W.; Cho, N.-J. Biotechnology Applications of Tethered Lipid Bilayer Membranes. Materials 2012, 5, 2637.
- Chadli, M.; Maniti, O.; Marquette, C.; Tillier, B.; Cortes, S.; Girard-Egrot, A. A new functional membrane protein microarray based on tethered phospholipid bilayers. Analyst 2018, 143, 2165–2173.
- Rebaud, S.; Maniti, O.; Girard-Egrot, A.P. Tethered bilayer lipid membranes (tBLMs): Interest and applications for biological membrane investigations. Biochimie 2014, 107 Pt A, 135–142.
- Koenig, B.W.; Krueger, S.; Orts, W.J.; Majkrzak, C.F.; Berk, N.F.; Silverton, J.V.; Gawrisch, K. Neutron Reflectivity and Atomic Force Microscopy Studies of a Lipid Bilayer in Water Adsorbed to the Surface of a Silicon Single Crystal. Langmuir 1996, 12, 1343–1350.
- Richter, R.P.; Bérat, R.; Brisson, A.R. Formation of Solid-Supported Lipid Bilayers: An Integrated View. Langmuir 2006, 22, 3497–3505.
- Keller, C.A.; Glasmästar, K.; Zhdanov, V.P.; Kasemo, B. Formation of Supported Membranes from Vesicles. Phys. Rev. Lett. 2000, 84, 5443–5446.
- Lind, T.K.; Cárdenas, M. Understanding the formation of supported lipid bilayers via vesicle fusion—A case that exemplifies the need for the complementary method approach (Review). Biointerphases 2016, 11, 020801.
- Johnson, S.J.; Bayerl, T.M.; McDermott, D.C.; Adam, G.W.; Rennie, A.R.; Thomas, R.K.; Sackmann, E. Structure of an adsorbed dimyristoylphosphatidylcholine bilayer measured with specular reflection of neutrons. Biophys. J. 1991, 59, 289–294.
- Tero, R. Substrate Effects on the Formation Process, Structure and Physicochemical Properties of Supported Lipid Bilayers. Materials 2012, 5, 2658–2680.
- Andersson, J.; Köper, I. Tethered and Polymer Supported Bilayer Lipid Membranes: Structure and Function. Membranes 2016, 6, 30.
- Groves, J.T.; Dustin, M.L. Supported planar bilayers in studies on immune cell adhesion and communication. J. Immunol. Methods 2003, 278, 19–32.
- Reviakine, I.; Brisson, A. Streptavidin 2D Crystals on Supported Phospholipid Bilayers: Toward Constructing Anchored Phospholipid Bilayers. Langmuir 2001, 17, 8293–8299.
- Milhiet, P.-E.; Giocondi, M.-C.; Baghdadi, O.; Ronzon, F.; Roux, B.; Le Grimellec, C. Spontaneous insertion and partitioning of alkaline phosphatase into model lipid rafts. EMBO Rep. 2002, 3, 485–490.
- Bouter, A.; Gounou, C.; Bérat, R.; Tan, S.; Gallois, B.; Granier, T.; d’Estaintot, B.L.; Pöschl, E.; Brachvogel, B.; Brisson, A.R. Annexin-A5 assembled into two-dimensional arrays promotes cell membrane repair. Nat. Commun. 2011, 2, 270.
- Heath, G.R.; Scheuring, S. High-speed AFM height spectroscopy reveals µs-dynamics of unlabeled biomolecules. Nat. Commun. 2018, 9, 4983.
- Melby, E.S.; Mensch, A.C.; Lohse, S.E.; Hu, D.; Orr, G.; Murphy, C.J.; Hamers, R.J.; Pedersen, J.A. Formation of supported lipid bilayers containing phase-segregated domains and their interaction with gold nanoparticles. Environ. Sci. Nano 2016, 3, 45–55.
- Waldie, S.; Lind, T.K.; Browning, K.; Moulin, M.; Haertlein, M.; Forsyth, V.T.; Luchini, A.; Strohmeier, G.A.; Pichler, H.; Maric, S.; et al. Localization of Cholesterol within Supported Lipid Bilayers Made of a Natural Extract of Tailor-Deuterated Phosphatidylcholine. Langmuir 2018, 34, 472–479.
- Waldie, S.; Moulin, M.; Porcar, L.; Pichler, H.; Strohmeier, G.A.; Skoda, M.; Forsyth, V.T.; Haertlein, M.; Maric, S.; Cárdenas, M. The Production of Matchout-Deuterated Cholesterol and the Study of Bilayer-Cholesterol Interactions. Sci. Rep. 2019, 9, 5118.
- Shilts, K.; Naumann, C.A. Tunable cell-surface mimetics as engineered cell substrates. Biochim. Biophys. Acta Biomembr. 2018, 1860, 2076–2093.
- Yu, C.-h.; Groves, J. Engineering supported membranes for cell biology. Med. Biol. Eng. Comput. 2010, 48, 955–963.
- Hartman, N.C.; Nye, J.A.; Groves, J.T. Cluster size regulates protein sorting in the immunological synapse. Proc. Natl. Acad. Sci. USA 2009, 106, 12729–12734.
- Torres, A.J.; Contento, R.L.; Gordo, S.; Wucherpfennig, K.W.; Love, J.C. Functional single-cell analysis of T-cell activation by supported lipid bilayer-tethered ligands on arrays of nanowells. Lab. Chip 2013, 13, 90–99.
- Groves, J.T.; Parthasarathy, R.; Forstner, M.B. Fluorescence Imaging of Membrane Dynamics. Annu. Rev. Biomed. Eng. 2008, 10, 311–338.
- Loose, M.; Schwille, P. Biomimetic membrane systems to study cellular organization. J. Struct. Biol. 2009, 168, 143–151.
- Cho, N.-J.; Frank, C.W.; Kasemo, B.; Hook, F. Quartz crystal microbalance with dissipation monitoring of supported lipid bilayers on various substrates. Nat. Protoc. 2010, 5, 1096–1106.
- Przybylo, M.; Sýkora, J.; Humpolíčková, J.; Benda, A.; Zan, A.; Hof, M. Lipid Diffusion in Giant Unilamellar Vesicles Is More than 2 Times Faster than in Supported Phospholipid Bilayers under Identical Conditions. Langmuir 2006, 22, 9096–9099.
- Macháň, R.; Hof, M. Lipid diffusion in planar membranes investigated by fluorescence correlation spectroscopy. Biochim. Biophys. Acta Biomembr. 2010, 1798, 1377–1391.
- Wu, H.-L.; Tong, Y.; Peng, Q.; Li, N.; Ye, S. Phase transition behaviors of the supported DPPC bilayer investigated by sum frequency generation (SFG) vibrational spectroscopy and atomic force microscopy (AFM). Phys. Chem. Chem. Phys. 2016, 18, 1411–1421.
- Sondhi, P.; Lingden, D.; Stine, K.J. Structure, Formation, and Biological Interactions of Supported Lipid Bilayers (SLB) Incorporating Lipopolysaccharide. Coatings 2020, 10, 981.
- Sinner, E.-K.; Ritz, S.; Naumann, R.; Schiller, S.; Knoll, W. Self-Assembled Tethered Bimolecular Lipid Membranes. In Advances in Clinical Chemistry; Gregory, S.M., Ed.; Elsevier: Amsterdam, The Netherlands, 2009; Volume 49, pp. 159–179.
- Köper, I.; Schiller, S.M.; Giess, F.; Naumann, R.; Knoll, W. Functional Tethered Bimolecular Lipid Membranes (tBLMs). In Advances in Planar Lipid Bilayers and Liposomes; Liu, A.L., Ed.; Academic Press: Cambridge, MA, USA, 2006; Volume 3, pp. 37–53.
- Tanaka, M.; Rosseti, F.F.; Kaufmann, S. Native supported membranes: Creation of two-dimensional cell membranes on polymer supports (Review). Biointerphases 2008, 3, FA12–FA16.
- Lind, T.K.; Wacklin, H.; Schiller, J.; Moulin, M.; Haertlein, M.; Pomorski, T.G.; Cárdenas, M. Formation and Characterization of Supported Lipid Bilayers Composed of Hydrogenated and Deuterated Escherichia coli Lipids. PLoS ONE 2015, 10, e0144671.
- Reimhult, E.; Kumar, K. Membrane biosensor platforms using nano- and microporous supports. Trends Biotechnol. 2008, 26, 82–89.
- Knoll, W.; Köper, I.; Naumann, R.; Sinner, E.-K. Tethered bimolecular lipid membranes—A novel model membrane platform. Electrochim. Acta 2008, 53, 6680–6689.
- Junghans, A.; Köper, I. Structural Analysis of Tethered Bilayer Lipid Membranes. Langmuir 2010, 26, 11035–11040.
- Liu, C.; Faller, R. Conformational, Dynamical. and Tensional Study of Tethered Bilayer Lipid Membranes in Coarse-Grained Molecular Simulations. Langmuir 2012, 28, 15907–15915.
- Montal, M.; Mueller, P. Formation of Bimolecular Membranes from Lipid Monolayers and a Study of Their Electrical Properties. Proc. Natl. Acad. Sci. USA 1972, 69, 3561–3566.
- Tien, H.T.; Ottova, A.L. The lipid bilayer concept and its experimental realization: From soap bubbles, kitchen sink, to bilayer lipid membranes. J. Membr. Sci. 2001, 189, 83–117.
- Berquand, A.; Mazeran, P.-E.; Pantigny, J.; Proux-Delrouyre, V.; Laval, J.-M.; Bourdillon, C. Two-Step Formation of Streptavidin-Supported Lipid Bilayers by PEG-Triggered Vesicle Fusion. Fluorescence and Atomic Force Microscopy Characterization†. Langmuir 2003, 19, 1700–1707.
- Jeuken, L.J.C.; Daskalakis, N.N.; Han, X.; Sheikh, K.; Erbe, A.; Bushby, R.J.; Evans, S.D. Phase separation in mixed self-assembled monolayers and its effect on biomimetic membranes. Sens. Actuators B 2007, 124, 501–509.
- Lee, B.K.; Lee, H.Y.; Kim, P.; Suh, K.Y.; Kawai, T. Nanoarrays of tethered lipid bilayer rafts on poly(vinyl alcohol) hydrogels. Lab. Chip 2009, 9, 132–139.
- Vockenroth, I.K.; Rossi, C.; Shah, M.R.; Köper, I. Formation of tethered bilayer lipid membranes probed by various surface sensitive techniques. Biointerphases 2009, 4, 19–26.
- Basit, H.; Van der Heyden, A.; Gondran, C.; Nysten, B.; Dumy, P.; Labbé, P. Tethered Bilayer Lipid Membranes on Mixed Self-Assembled Monolayers of a Novel Anchoring Thiol: Impact of the Anchoring Thiol Density on Bilayer Formation. Langmuir 2011, 27, 14317–14328.
- Bronder, A.M.; Bieker, A.; Elter, S.; Etzkorn, M.; Häussinger, D.; Oesterhelt, F. Oriented Membrane Protein Reconstitution into Tethered Lipid Membranes for AFM Force Spectroscopy. Biophys. J. 2016, 111, 1925–1934.
- Zhou, W.; Burke, P.J. Versatile Bottom-Up Synthesis of Tethered Bilayer Lipid Membranes on Nanoelectronic Biosensor Devices. ACS Appl. Mater. Interfaces 2017, 9, 14618–14632.
- Shenoy, S.; Moldovan, R.; Fitzpatrick, J.; Vanderah, D.J.; Deserno, M.; Lösche, M. In-plane homogeneity and lipid dynamics in tethered bilayer lipid membranes (tBLMs). Soft Matter 2010, 6, 1263–1274.
- Eicher-Lorka, O.; Charkova, T.; Matijoška, A.; Kuodis, Z.; Urbelis, G.; Penkauskas, T.; Mickevičius, M.; Bulovas, A.; Valinčius, G. Cholesterol-based tethers and markers for model membranes investigation. Chem. Phys. Lipids 2016, 195, 71–86.
- Naumann, C.A.; Prucker, O.; Lehmann, T.; Rühe, J.; Knoll, W.; Frank, C.W. The Polymer-Supported Phospholipid Bilayer: Tethering as a New Approach to Substrate−Membrane Stabilization. Biomacromolecules 2002, 3, 27–35.
- Munro, J.C.; Frank, C.W. In Situ Formation and Characterization of Poly(ethylene glycol)-Supported Lipid Bilayers on Gold Surfaces. Langmuir 2004, 20, 10567–10575.
- Liu, H.-Y.; Chen, W.-L.; Ober, C.K.; Daniel, S. Biologically Complex Planar Cell Plasma Membranes Supported on Polyelectrolyte Cushions Enhance Transmembrane Protein Mobility and Retain Native Orientation. Langmuir 2018, 34, 1061–1072.
- Roder, F.; Waichman, S.; Paterok, D.; Schubert, R.; Richter, C.; Liedberg, B.; Piehler, J. Reconstitution of Membrane Proteins into Polymer-Supported Membranes for Probing Diffusion and Interactions by Single Molecule Techniques. Anal. Chem. 2011, 83, 6792–6799.
- McGillivray, D.J.; Valincius, G.; Vanderah, D.J.; Febo-Ayala, W.; Woodward, J.T.; Heinrich, F.; Kasianowicz, J.J.; Lösche, M. Molecular-scale structural and functional characterization of sparsely tethered bilayer lipid membranes. Biointerphases 2007, 2, 21–33.
- Vockenroth, I.K.; Ohm, C.; Robertson, J.W.F.; McGillivraya, D.J.; Lösche, M.; Köper, I. Stable insulating tethered bilayer lipid membranes. Biointerphases 2008, 3, FA68–FA73.
- Budvytyte, R.; Valincius, G.; Niaura, G.; Voiciuk, V.; Mickevicius, M.; Chapman, H.; Goh, H.-Z.; Shekhar, P.; Heinrich, F.; Shenoy, S.; et al. Structure and Properties of Tethered Bilayer Lipid Membranes with Unsaturated Anchor Molecules. Langmuir 2013, 29, 8645–8656.
- Hertrich, S.; Stetter, F.; Rühm, A.; Hugel, T.; Nickel, B. Highly Hydrated Deformable Polyethylene Glycol-Tethered Lipid Bilayers. Langmuir 2014, 30, 9442–9447.
- Yap, T.L.; Jiang, Z.; Heinrich, F.; Gruschus, J.M.; Pfefferkorn, C.M.; Barros, M.; Curtis, J.E.; Sidransky, E.; Lee, J.C. Structural features of membrane-bound glucocerebrosidase and α-synuclein probed by neutron reflectometry and fluorescence spectroscopy. J. Biol. Chem. 2015, 290, 744–754.
- Maccarini, M.; Watkins, E.B.; Stidder, B.; Alcaraz, J.-P.; Cornell, B.A.; Martin, D.K. Nanostructural determination of a lipid bilayer tethered to a gold substrate. Eur. Phys. J. E 2016, 39, 123.
- Cranfield, C.G.; Berry, T.; Holt, S.A.; Hossain, K.R.; Le Brun, A.P.; Carne, S.; Al Khamici, H.; Coster, H.; Valenzuela, S.M.; Cornell, B. Evidence of the Key Role of H3O+ in Phospholipid Membrane Morphology. Langmuir 2016, 32, 10725–10734.
- Andersson, J.; Knobloch, J.J.; Perkins, M.V.; Holt, S.A.; Köper, I. Synthesis and Characterization of Novel Anchorlipids for Tethered Bilayer Lipid Membranes. Langmuir 2017, 33, 4444–4451.
- Alharbi, A.R.M.; Andersson, J.M.; Köper, I.; Andersson, G.G. Investigating the Structure of Self-Assembled Monolayers Related to Biological Cell Membranes. Langmuir 2019, 35, 14213–14221.
- Squillace, O.; Perrault, T.; Gorczynska, M.; Caruana, A.; Bajorek, A.; Brotons, G. Design of tethered bilayer lipid membranes, using wet chemistry via aryldiazonium sulfonic acid spontaneous grafting on silicon and chrome. Colloids Surf. B 2021, 197, 111427.
- Atanasov, V.; Knorr, N.; Duran, R.S.; Ingebrandt, S.; Offenhäusser, A.; Knoll, W.; Köper, I. Membrane on a Chip: A Functional Tethered Lipid Bilayer Membrane on Silicon Oxide Surfaces. Biophys. J. 2005, 89, 1780–1788.
- Leitch, J.; Kunze, J.; Goddard, J.D.; Schwan, A.L.; Faragher, R.J.; Naumann, R.; Knoll, W.; Dutcher, J.R.; Lipkowski, J. In Situ PM-IRRAS Studies of an Archaea Analogue Thiolipid Assembled on a Au(111) Electrode Surface. Langmuir 2009, 25, 10354–10363.
- Su, Z.; Ran, X.; Leitch, J.J.; Schwan, A.L.; Faragher, R.; Lipkowski, J. How Valinomycin Ionophores Enter and Transport K+ across Model Lipid Bilayer Membranes. Langmuir 2019, 35, 16935–16943.
- Ataka, K.; Giess, F.; Knoll, W.; Naumann, R.; Haber-Pohlmeier, S.; Richter, B.; Heberle, J. Oriented Attachment and Membrane Reconstitution of His-Tagged Cytochrome c Oxidase to a Gold Electrode: In Situ Monitoring by Surface-Enhanced Infrared Absorption Spectroscopy. J. Am. Chem. Soc. 2004, 126, 16199–16206.
- Kozuch, J.; Weichbrodt, C.; Millo, D.; Giller, K.; Becker, S.; Hildebrandt, P.; Steinem, C. Voltage-dependent structural changes of the membrane-bound anion channel hVDAC1 probed by SEIRA and electrochemical impedance spectroscopy. Phys. Chem. Chem. Phys. 2014, 16, 9546–9555.
- Wiebalck, S.; Kozuch, J.; Forbrig, E.; Tzschucke, C.C.; Jeuken, L.J.C.; Hildebrandt, P. Monitoring the Transmembrane Proton Gradient Generated by Cytochrome bo3 in Tethered Bilayer Lipid Membranes Using SEIRA Spectroscopy. J. Phys. Chem. B 2016, 120, 2249–2256.
- Forbrig, E.; Staffa, J.K.; Salewski, J.; Mroginski, M.A.; Hildebrandt, P.; Kozuch, J. Monitoring the Orientational Changes of Alamethicin during Incorporation into Bilayer Lipid Membranes. Langmuir 2018, 34, 2373–2385.
- Schmidt, E.K.; Liebermann, T.; Kreiter, M.; Jonczyk, A.; Naumann, R.; Offenhäusser, A.; Neumann, E.; Kukol, A.; Maelicke, A.; Knoll, W. Incorporation of the acetylcholine receptor dimer from Torpedo californica in a peptide supported lipid membrane investigated by surface plasmon and fluorescence spectroscopy. Biosens. Bioelectron. 1998, 13, 585–591.
- Giess, F.; Friedrich, M.G.; Heberle, J.; Naumann, R.L.; Knoll, W. The Protein-Tethered Lipid Bilayer: A Novel Mimic of the Biological Membrane. Biophys. J. 2004, 87, 3213–3220.
- Wiltschi, B.; Knoll, W.; Sinner, E.-K. Binding assays with artificial tethered membranes using surface plasmon resonance. Methods 2006, 39, 134–146.
- Becucci, L.; Moncelli, M.R.; Naumann, R.; Guidelli, R. Potassium Ion Transport by Valinomycin across a Hg-Supported Lipid Bilayer. J. Am. Chem. Soc. 2005, 127, 13316–13323.
- Naumann, R.; Baumgart, T.; Gräber, P.; Jonczyk, A.; Offenhäusser, A.; Knoll, W. Proton transport through a peptide-tethered bilayer lipid membrane by the H+−ATP synthase from chloroplasts measured by impedance spectroscopy. Biosens. Bioelectron. 2002, 17, 25–34.
- Krishna, G.; Schulte, J.; Cornell, B.A.; Pace, R.J.; Osman, P.D. Tethered Bilayer Membranes Containing Ionic Reservoirs: Selectivity and Conductance. Langmuir 2003, 19, 2294–2305.
- Cranfield, C.G.; Bettler, T.; Cornell, B. Nanoscale Ion Sequestration To Determine the Polarity Selectivity of Ion Conductance in Carriers and Channels. Langmuir 2015, 31, 292–298.
- Proux-Delrouyre, V.; Elie, C.; Laval, J.-M.; Moiroux, J.; Bourdillon, C. Formation of Tethered and Streptavidin-Supported Lipid Bilayers on a Microporous Electrode for the Reconstitution of Membranes of Large Surface Area. Langmuir 2002, 18, 3263–3272.
- Jeuken, L.J.C.; Connell, S.D.; Henderson, P.J.F.; Gennis, R.B.; Evans, S.D.; Bushby, R.J. Redox Enzymes in Tethered Membranes. J. Am. Chem. Soc. 2006, 128, 1711–1716.
- Friedrich, M.G.; Robertson, J.W.F.; Walz, D.; Knoll, W.; Naumann, R.L.C. Electronic Wiring of a Multi-Redox Site Membrane Protein in a Biomimetic Surface Architecture. Biophys. J. 2008, 94, 3698–3705.
- Jeuken, L.J.C. Electrodes for integral membrane enzymes. Nat. Prod. Rep. 2009, 26, 1234–1240.
- Nowak, C.; Schach, D.; Gebert, J.; Grosserueschkamp, M.; Gennis, R.B.; Ferguson-Miller, S.; Knoll, W.; Walz, D.; Naumann, R.L.C. Oriented immobilization and electron transfer to the cytochrome c oxidase. J. Solid State Electrochem. 2011, 15, 105–114.
- Becucci, L.; Guidelli, R. Can gramicidin ion channel affect the dipole potential of neighboring phospholipid headgroups? Bioelectrochemistry 2015, 106, 343–352.
- McGillivray, D.J.; Valincius, G.; Heinrich, F.; Robertson, J.W.F.; Vanderah, D.J.; Febo-Ayala, W.; Ignatjev, I.; Lösche, M.; Kasianowicz, J.J. Structure of Functional Staphylococcus aureus α-Hemolysin Channels in Tethered Bilayer Lipid Membranes. Biophys. J. 2009, 96, 1547–1553.
- Andersson, J.; Köper, I.; Knoll, W. Tethered Membrane Architectures—Design and Applications. Front. Mater. 2018, 5.
- Rossi, C.; Chopineau, J. Biomimetic tethered lipid membranes designed for membrane-protein interaction studies. Eur. Biophys. J. 2007, 36, 955–965.
