Biliary tract cancer (BTC) is often refractory to conventional therapeutics and is difficult to diagnose in the early stages. Implantation-based models have recently drawn attention for their convenience, flexibility, and scalability.
- genetically engineered mouse
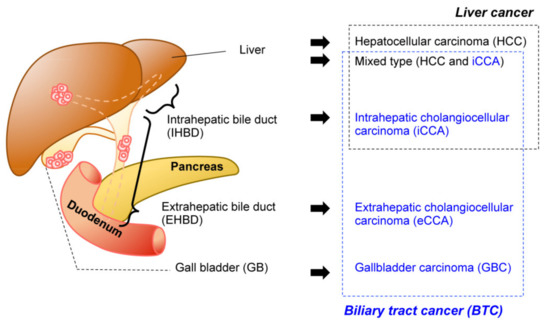
- biliary tract cancer
- organoid
- orthotopic model
- nude mouse
- syngeneic
- hydrodynamic injection
- implantation
1. Introduction
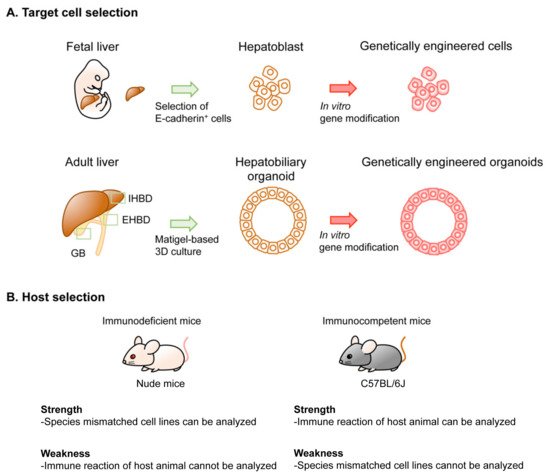
2. Technical Overview of Genetic Mouse Models of BTC
|
Driver Oncogenes |
Genotype of Organoids * |
Methods for Genetic Engineering ** |
Host |
Implantation |
Ref. |
|
|---|---|---|---|---|---|---|
|
Oncogenes |
TSGs |
|||||
|
HCC (Hepatocellular Carcinoma) from Liver Organoid |
||||||
|
cMyc |
WT |
cDNA (R) |
shRNA (R); Trp53 and CRISPR/Cas9 (T); Apc |
C57BL/6J |
liver |
[18] |
|
iCCA (Intrahepatic Cholangiocelular Carcinoma) from Liver Organoid |
||||||
|
KrasG12D |
KrasLSL-G12D/+ |
Cre (L) |
shRNA (L): Cdkn2a and/or Pten, Trp53, Apc |
Nude |
s.c. |
[19] |
|
KrasLSL-G12D/+; Trp53flox/flox |
N.T. |
|||||
|
Pik3caH1047R |
Pik3caH1047R |
shRNA (L): Cdkn2a, Pten, |
||||
|
Rosa26- Pik3caH1047R; Trp53flox/flox |
N.T. |
|||||
|
FGFR2-AHCYL1 |
WT |
cDNA(R) |
shRNA (L): Cdkn2a and/or Pten |
|||
|
KrasG12D |
KrasLSL-G12D/+; Trp53flox/flox (outbred) |
Cre (R) |
N.T. or shRNA (R): Pten |
NSG |
s.c. or liver |
[18] |
|
KrasLSL-G12D/+ |
Cre (T) |
CRISPR/Cas9 (T): Pten and Trp53 |
C57BL/6J |
liver |
||
|
FGFR2-BICC1, -MGEA5, -TACC3 |
Trp53−/− |
cDNA (R) |
N.T. |
NOD-SCID |
s.c. or liver |
[20] |
|
KRASG12V |
Cdkn2a−/− |
cDNA (R) |
C57BL/6J |
s.c., liver, or kidney |
[21] |
|
|
eCCA (Extrahepatic Cholangiocelular Carcinoma) from CBD Organoid |
||||||
|
KrasG12D |
KrasLSL-G12D; Tgfbr2flox/flox; Cdh1flox/flox |
Cre (L) |
N.T. |
Nude or C57BL/6J |
s.c. |
[22] |
|
KRASG12V |
Cdkn2a−/− |
cDNA (R) |
N.T. |
C57BL/6J |
s.c., liver, or kidney |
[21] |
|
GBC (Gallbladder Carcinoma) from GB Organoid |
||||||
|
KrasG12D |
KrasLSL-G12D |
Cre (L) |
shRNA (L): Cdkn2a, Pten |
Nude |
s.c. |
[19] |
|
KrasLSL-G12D/+; Trp53flox/flox |
N.T. |
|||||
|
KRASG12V |
Cdkn2a−/− |
cDNA (R) |
N.T. |
C57BL/6J |
s.c., liver, or kidney |
[21] |
|
KrasG12D |
WT |
Cre (T) |
CRISPR/Cas9 (T): Pten and Trp53 |
C57BL/6J |
s.c. or GB |
[23] |
|
ERBB2S310F or V777L |
cDNA (R) |
CRISPR/Cas9 (T): Trp53 |
NSG |
s.c. |
||
|
Rosa26- Pik3caH1047R; Trp53flox/flox |
Cre (L) |
N.T. |
Nude |
s.c. |
[24] |
|
|
KrasG12D |
KrasLSL-G12D |
CRISPR/Cas9 (T): Trp53, p19Arf, Smad4 |
C57BL/6J |
GB via s.c. |
||
For those models that developed cancer, pre-cancerous lesion and cystic lesions were listed. Note that liver organoid-derived tumor are either HCC or CCA (adenocarcinoma), but not mixed type liver cancer * C57BL/6J background unless otherwise indicated. ** L, lentivirus; R, retrovirus; T, transfection; N.T., not tested. Abbreviations: TSG, tumor suppressor genes; s.c., subcutis; WT, wildtype; CBD, common bile duct; GB, gallbladder.
3. In Vivo GEM Model of BTC
4. Implantation-Based Hybrid Mouse Model of BTC
This entry is adapted from the peer-reviewed paper 10.3390/cancers13102292
References
- Banales, J.M.; Marin, J.J.G.; Lamarca, A.; Rodrigues, P.M.; Khan, S.A.; Roberts, L.R.; Cardinale, V.; Carpino, G.; Andersen, J.B.; Braconi, C.; et al. Cholangiocarcinoma 2020: The next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 557–588.
- Khan, S.A.; Tavolari, S.; Brandi, G. Cholangiocarcinoma: Epidemiology and risk factors. Liver Int. 2019, 39, 19–31.
- Bertuccio, P.; Malvezzi, M.; Carioli, G.; Hashim, D.; Boffetta, P.; El-Serag, H.B.; La Vecchia, C.; Negri, E. Global trends in mortality from intrahepatic and extrahepatic cholangiocarcinoma. J. Hepatol. 2019, 71, 104–114.
- Florio, A.A.; Ferlay, J.; Znaor, A.; Ruggieri, D.; Alvarez, C.S.; Laversanne, M.; Bray, F.; McGlynn, K.A.; Petrick, J.L. Global trends in intrahepatic and extrahepatic cholangiocarcinoma incidence from 1993 to 2012. Cancer 2020, 126, 2666–2678.
- Ainechi, S.; Lee, H. Updates on Precancerous Lesions of the Biliary Tract: Biliary Precancerous Lesion. Arch. Pathol. Lab. Med. 2016, 140, 1285–1289.
- Kendall, T.; Verheij, J.; Gaudio, E.; Evert, M.; Guido, M.; Goeppert, B.; Carpino, G. Anatomical, histomorphological and molecular classification of cholangiocarcinoma. Liver Int. 2019, 39, 7–18.
- Chan-On, W.; Nairismagi, M.L.; Ong, C.K.; Lim, W.K.; Dima, S.; Pairojkul, C.; Lim, K.H.; McPherson, J.R.; Cutcutache, I.; Heng, H.L.; et al. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat. Genet. 2013, 45, 1474–1478.
- Jiao, Y.; Pawlik, T.M.; Anders, R.A.; Selaru, F.M.; Streppel, M.M.; Lucas, D.J.; Niknafs, N.; Guthrie, V.B.; Maitra, A.; Argani, P.; et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat. Genet. 2013, 45, 1470–1473.
- Jusakul, A.; Cutcutache, I.; Yong, C.H.; Lim, J.Q.; Huang, M.N.; Padmanabhan, N.; Nellore, V.; Kongpetch, S.; Ng, A.W.T.; Ng, L.M.; et al. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov. 2017, 7, 1116–1135.
- Li, M.; Zhang, Z.; Li, X.; Ye, J.; Wu, X.; Tan, Z.; Liu, C.; Shen, B.; Wang, X.A.; Wu, W.; et al. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat. Genet. 2014, 46, 872–876.
- Ong, C.K.; Subimerb, C.; Pairojkul, C.; Wongkham, S.; Cutcutache, I.; Yu, W.; McPherson, J.R.; Allen, G.E.; Ng, C.C.; Wong, B.H.; et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat. Genet. 2012, 44, 690–693.
- Sia, D.; Losic, B.; Moeini, A.; Cabellos, L.; Hao, K.; Revill, K.; Bonal, D.; Miltiadous, O.; Zhang, Z.; Hoshida, Y.; et al. Massive parallel sequencing uncovers actionable FGFR2-PPHLN1 fusion and ARAF mutations in intrahepatic cholangiocarcinoma. Nat. Commun. 2015, 6, 6087.
- Zou, S.; Li, J.; Zhou, H.; Frech, C.; Jiang, X.; Chu, J.S.; Zhao, X.; Li, Y.; Li, Q.; Wang, H.; et al. Mutational landscape of intrahepatic cholangiocarcinoma. Nat. Commun. 2014, 5, 5696.
- Nakamura, H.; Arai, Y.; Totoki, Y.; Shirota, T.; Elzawahry, A.; Kato, M.; Hama, N.; Hosoda, F.; Urushidate, T.; Ohashi, S.; et al. Genomic spectra of biliary tract cancer. Nat. Genet. 2015, 47, 1003–1010.
- Massa, A.; Varamo, C.; Vita, F.; Tavolari, S.; Peraldo-Neia, C.; Brandi, G.; Rizzo, A.; Cavalloni, G.; Aglietta, M. Evolution of the Experimental Models of Cholangiocarcinoma. Cancers 2020, 12, 2308.
- Leiting, J.L.; Murphy, S.J.; Bergquist, J.R.; Hernandez, M.C.; Ivanics, T.; Abdelrahman, A.M.; Yang, L.; Lynch, I.; Smadbeck, J.B.; Cleary, S.P.; et al. Biliary tract cancer patient-derived xenografts: Surgeon impact on individualized medicine. JHEP Rep. 2020, 2, 100068.
- Waddell, S.H.; Boulter, L. Developing models of cholangiocarcinoma to close the translational gap in cancer research. Expert Opin. Investig. Drugs 2021, 30, 439–450.
- Saborowski, A.; Wolff, K.; Spielberg, S.; Beer, B.; Hartleben, B.; Erlangga, Z.; Becker, D.; Dow, L.E.; Marhenke, S.; Woller, N.; et al. Murine Liver Organoids as a Genetically Flexible System to Study Liver Cancer In Vivo and In Vitro. Hepatol. Commun. 2019, 3, 423–436.
- Ochiai, M.; Yoshihara, Y.; Maru, Y.; Tetsuya, M.; Izumiya, M.; Imai, T.; Hippo, Y. Kras-driven heterotopic tumor development from hepatobiliary organoids. Carcinogenesis 2019, 40, 1142–1152.
- Cristinziano, G.; Porru, M.; Lamberti, D.; Buglioni, S.; Rollo, F.; Amoreo, C.A.; Manni, I.; Giannarelli, D.; Cristofoletti, C.; Russo, G.; et al. FGFR2 fusion proteins drive oncogenic transformation of mouse liver organoids towards cholangiocarcinoma. J. Hepatol. 2021.
- Kasuga, A.; Semba, T.; Sato, R.; Nobusue, H.; Sugihara, E.; Takaishi, H.; Kanai, T.; Saya, H.; Arima, Y. Oncogenic KRAS-expressing organoids with biliary epithelial stem cell properties give rise to biliary tract cancer in mice. Cancer Sci. 2021, 112, 1822–1838.
- Nakagawa, H.; Suzuki, N.; Hirata, Y.; Hikiba, Y.; Hayakawa, Y.; Kinoshita, H.; Ihara, S.; Uchino, K.; Nishikawa, Y.; Ijichi, H.; et al. Biliary epithelial injury-induced regenerative response by IL-33 promotes cholangiocarcinogenesis from peribiliary glands. Proc. Natl. Acad. Sci. USA 2017, 114, E3806–E3815.
- Erlangga, Z.; Wolff, K.; Poth, T.; Peltzer, A.; Nahnsen, S.; Spielberg, S.; Timrott, K.; Woller, N.; Kuhnel, F.; Manns, M.P.; et al. Potent Antitumor Activity of Liposomal Irinotecan in an Organoid- and CRISPR-Cas9-Based Murine Model of Gallbladder Cancer. Cancers 2019, 11, 1904.
- Kato, S.; Fushimi, K.; Yabuki, Y.; Maru, Y.; Hasegawa, S.; Matsuura, T.; Kurotaki, D.; Suzuki, A.; Kobayashi, N.; Yoneda, M.; et al. Precision modeling of gall bladder cancer patients in mice based on orthotopic implantation of organoid-derived tumor buds. Oncogenesis 2021, 10, 33.
- Xu, X.; Kobayashi, S.; Qiao, W.; Li, C.; Xiao, C.; Radaeva, S.; Stiles, B.; Wang, R.H.; Ohara, N.; Yoshino, T.; et al. Induction of intrahepatic cholangiocellular carcinoma by liver-specific disruption of Smad4 and Pten in mice. J. Clin. Investig. 2006, 116, 1843–1852.
- Ikenoue, T.; Terakado, Y.; Nakagawa, H.; Hikiba, Y.; Fujii, T.; Matsubara, D.; Noguchi, R.; Zhu, C.; Yamamoto, K.; Kudo, Y.; et al. A novel mouse model of intrahepatic cholangiocarcinoma induced by liver-specific Kras activation and Pten deletion. Sci. Rep. 2016, 6, 23899.
- Galicia, V.A.; He, L.; Dang, H.; Kanel, G.; Vendryes, C.; French, B.A.; Zeng, N.; Bayan, J.A.; Ding, W.; Wang, K.S.; et al. Expansion of hepatic tumor progenitor cells in Pten-null mice requires liver injury and is reversed by loss of AKT2. Gastroenterology 2010, 139, 2170–2182.
- Hill, M.A.; Alexander, W.B.; Guo, B.; Kato, Y.; Patra, K.; O’Dell, M.R.; McCall, M.N.; Whitney-Miller, C.L.; Bardeesy, N.; Hezel, A.F. Kras and Tp53 Mutations Cause Cholangiocyte- and Hepatocyte-Derived Cholangiocarcinoma. Cancer Res. 2018, 78, 4445–4451.
- Ikenoue, T.; Terakado, Y.; Zhu, C.; Liu, X.; Ohsugi, T.; Matsubara, D.; Fujii, T.; Kakuta, S.; Kubo, S.; Shibata, T.; et al. Establishment and analysis of a novel mouse line carrying a conditional knockin allele of a cancer-specific FBXW7 mutation. Sci. Rep. 2018, 8, 2021.
- Lee, K.P.; Lee, J.H.; Kim, T.S.; Kim, T.H.; Park, H.D.; Byun, J.S.; Kim, M.C.; Jeong, W.I.; Calvisi, D.F.; Kim, J.M.; et al. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 8248–8253.
- Lin, Y.K.; Fang, Z.; Jiang, T.Y.; Wan, Z.H.; Pan, Y.F.; Ma, Y.H.; Shi, Y.Y.; Tan, Y.X.; Dong, L.W.; Zhang, Y.J.; et al. Combination of Kras activation and PTEN deletion contributes to murine hepatopancreatic ductal malignancy. Cancer Lett. 2018, 421, 161–169.
- O’Dell, M.R.; Huang, J.L.; Whitney-Miller, C.L.; Deshpande, V.; Rothberg, P.; Grose, V.; Rossi, R.M.; Zhu, A.X.; Land, H.; Bardeesy, N.; et al. Kras(G12D) and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer Res. 2012, 72, 1557–1567.
- Saha, S.K.; Parachoniak, C.A.; Ghanta, K.S.; Fitamant, J.; Ross, K.N.; Najem, M.S.; Gurumurthy, S.; Akbay, E.A.; Sia, D.; Cornella, H.; et al. Mutant IDH inhibits HNF-4alpha to block hepatocyte differentiation and promote biliary cancer. Nature 2014, 513, 110–114.
- Zender, S.; Nickeleit, I.; Wuestefeld, T.; Sorensen, I.; Dauch, D.; Bozko, P.; El-Khatib, M.; Geffers, R.; Bektas, H.; Manns, M.P.; et al. A critical role for notch signaling in the formation of cholangiocellular carcinomas. Cancer Cell 2013, 23, 784–795.
- Means, A.L.; Xu, Y.; Zhao, A.; Ray, K.C.; Gu, G. A CK19(CreERT) knockin mouse line allows for conditional DNA recombination in epithelial cells in multiple endodermal organs. Genesis 2008, 46, 318–323.
- Chung, W.C.; Wang, J.; Zhou, Y.; Xu, K. Kras(G12D) upregulates Notch signaling to induce gallbladder tumorigenesis in mice. Oncoscience 2017, 4, 131–138.
- Kiguchi, K.; Bol, D.; Carbajal, S.; Beltran, L.; Moats, S.; Chan, K.; Jorcano, J.; DiGiovanni, J. Constitutive expression of erbB2 in epidermis of transgenic mice results in epidermal hyperproliferation and spontaneous skin tumor development. Oncogene 2000, 19, 4243–4254.
- Kiguchi, K.; Carbajal, S.; Chan, K.; Beltran, L.; Ruffino, L.; Shen, J.; Matsumoto, T.; Yoshimi, N.; DiGiovanni, J. Constitutive expression of ErbB-2 in gallbladder epithelium results in development of adenocarcinoma. Cancer Res. 2001, 61, 6971–6976.
- Onuma, K.; Ochiai, M.; Orihashi, K.; Takahashi, M.; Imai, T.; Nakagama, H.; Hippo, Y. Genetic reconstitution of tumorigenesis in primary intestinal cells. Proc. Natl. Acad. Sci. USA 2013, 110, 11127–11132.
- Maru, Y.; Onuma, K.; Ochiai, M.; Imai, T.; Hippo, Y. Shortcuts to intestinal carcinogenesis by genetic engineering in organoids. Cancer Sci. 2019, 110, 858–866.
- Matsuura, T.; Maru, Y.; Izumiya, M.; Hoshi, D.; Kato, S.; Ochiai, M.; Hori, M.; Yamamoto, S.; Tatsuno, K.; Imai, T.; et al. Organoid-based ex vivo reconstitution of Kras-driven pancreatic ductal carcinogenesis. Carcinogenesis 2020, 41, 490–501.
- Sato, T.; Morita, M.; Tanaka, R.; Inoue, Y.; Nomura, M.; Sakamoto, Y.; Miura, K.; Ito, S.; Sato, I.; Tanaka, N.; et al. Ex vivo model of non-small cell lung cancer using mouse lung epithelial cells. Oncol. Lett. 2017, 14, 6863–6868.
- Naruse, M.; Masui, R.; Ochiai, M.; Maru, Y.; Hippo, Y.; Imai, T. An organoid-based carcinogenesis model induced by in vitro chemical treatment. Carcinogenesis 2020, 41, 1444–1453.
