Exosomes are membrane-bound nanovesicles that are typically 30–150 nm in size with various bioactive molecules. They are typically generated by first endocytosing various transmembrane proteins into endosomes within the cell, which are then sorted and form intraluminal vesicles. These vesicles are then released as the endosome merges with the cell membrane and releases its contents outside of the cell. Tetraspanins (CD9, CD63, CD81) are one of the most common proteins expressed on the surface of exosomes and are often used as exosome-specific markers. These proteins have been shown to interact with different proteins such as integrins and major histocompatibility complexes (MHC). Exosomes commonly act as carriers of genetic and proteomic information, and are therefore vital in intercellular communication. In its role as a cellular messenger, exosomes have been implicated in promoting cancer; because of this, they are also being investigated as potential therapeutic targets and delivery vehicles.
- exosome
- tumor microenvironment
- miRNAs
- lncRNAs
1. Role of Exosomes in Cancer-Associated Microenvironment
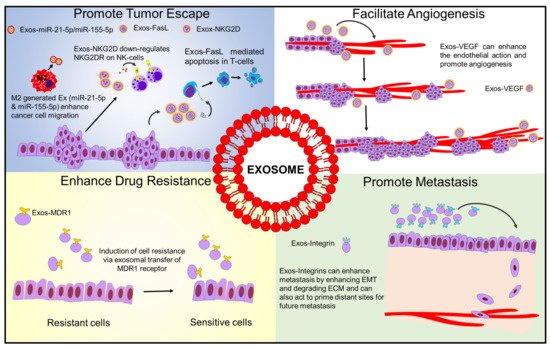
1.1. Exosome-Mediated Immune Evasion
1.2. Exosomes Enhance Cancer Progression and Metastasis
1.3. Exosomes and Drug Resistance
1.4. Exosomes Promoting Angiogenesis
1.5. Exosomal lncRNAs in Tumorigenesis
2. Common Isolation Methods of Exosomes
2.1. Ultracentrifugation
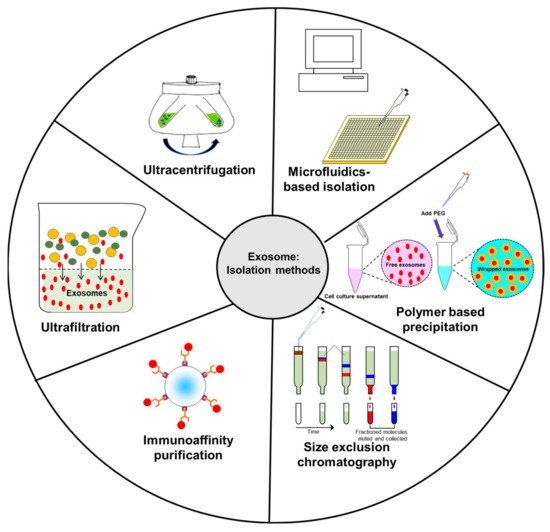
2.2. Ultrafiltration
2.3. Immunoaffinity Capture
2.4. Size Exclusion Chromatography (SEC)
2.5. Polymer-Based Precipitation
2.6. Microfluidics-Based Isolation
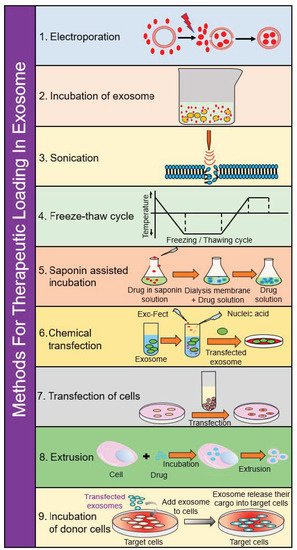
3. Methods for Therapeutic Loading in Exosomes
3.1. Electroporation
3.2. Incubation of Exosomes
3.3. Incubation of Donor Cells
3.4. Saponin-Assisted Incubation
3.5. Sonication
3.6. Extrusion
3.7. Freeze–Thaw Cycling
3.8. Chemical Transfection
3.9. Transfection of Cells
4. Use of Exosomes in Targeted Delivery
| Source of Exosomes | Cargo | Cancer Type | Key Findings | Reference Number |
|---|---|---|---|---|
| 293T cells | 5-FU, miR-21i | Colorectal | Cell cycle arrest, reduced proliferation, increased apoptosis; reduced cancer growth in vivo with minimal toxicity | [129] |
| Bone marrow mesenchymal stem cells | Galectin-9 siRNA and oxaliplatin | PDAC | Enhanced drug uptake, reduction of M2-like macrophages and enhanced anti-tumor immunity, greater tumor reduction in vivo | [119] |
| Autologous pancreatic cancer cells (Panc-1) and heterologous lung cancer cells (A549) | Gemcitabine | Pancreatic | Enhanced uptake of autologous exosomes; reduced tumor growth compared to free gemcitabine; enhanced survival in vivo | [130] |
| MDA-MB-231 and HT-29 cells | miRNA 126 | Non-small lung cell cancer | Enhanced uptake of MDA-MB-231 exosomes; reduction of proliferation and migration in vitro; reduced metastatic nodules in vivo with minimal toxicity | [131] |
| HeLa and L02 cells; exosomes isolated from blood cancer patient | Doxorubicin, silver nanoclusters and DNA | Cervical and blood cancers | Enhanced uptake of drug-loaded exosomes; superior theranostic capability compared to free silver nanoclusters | [118] |
| Cow milk and Caco-2 cells | Curcumin | Colorectal | Superior uptake noted with Caco-2-derived exosomes compared to milk-derived exosomes; enhanced therapeutic effect of CUR compared to free drug | [123] |
| Cow milk | Hyaluronan and doxorubicin | Breast, lung, kidney | Enhanced uptake into CD44 overexpressing cells, with superior cytotoxicity to free drug | [122] |
| Cow milk | Paclitaxel | Lung | Sustained release seen up to 48 h in vitro; significant growth inhibition after oral administration in vivo | [124] |
| Normal fibroblast-like mesenchymal cells | siRNA specific to oncogenic KRAS | Pancreatic | Superior KRAS targeting compared to liposomes; enhanced tumor suppression and survival in vivo | [126] |
| Umbilical cord macrophages | Cisplatin | Ovarian | Enhanced therapeutic effect of exosomal cisplatin in vitro against resistant and sensitive cell lines | [132] |
| Dental pulp mesenchymal stem cells | miR-34a | Breast | Downregulation of cancerous phenotype in vitro | [133] |
| Adipose tissue-derived mesenchymal stem cells | miR-199a | Hepatocellular carcinoma | Sensitized cancer cells to doxorubicin therapy both in vitro and in vivo | [134] |
| Mesenchymal stem cells | Doxorubicin | Colorectal | Significantly enhanced suppression of tumor growth in vivo compared to free doxorubicin | [135] |
| HEK293 cells | miR-34a | Pancreatic | Induced apoptosis in cancer cells in vitro; significant suppression of tumor growth in vivo | [136] |
| Bone marrow mesenchymal stem cells | miR-193a | Non-small cell lung cancer | Suppressed colony formation, proliferation, and invasion in vitro; reduced tumor volume in vivo | [137] |
5. Clinical Use of Exosomes: Progress and Promise
This entry is adapted from the peer-reviewed paper 10.3390/ijms22105278
