Taste sensation and regulation are highly conserved in insects and mammals. Research conducted over recent decades has yielded major advances in our understanding of the molecular mechanisms underlying the taste sensors for a variety of taste sensations and the processes underlying the regulation of ingestion depending on our internal state. Salt (NaCl) is an essential ingested nutrient. The regulation of internal sodium concentrations for physiological processes, including neuronal activity, fluid volume, acid-base balance, and muscle contraction, are extremely important issues in animal health. Both mammals and flies detect low and high NaCl concentrations as attractive and aversive tastants, respectively. These attractive or aversive behaviors can be modulated by the internal nutrient state. However, the differential encoding of the tastes underlying low and high salt concentrations in the brain remains unclear.
- Salt sensation
- salt coding
- taste perception
- salt appetite
1. Introduction
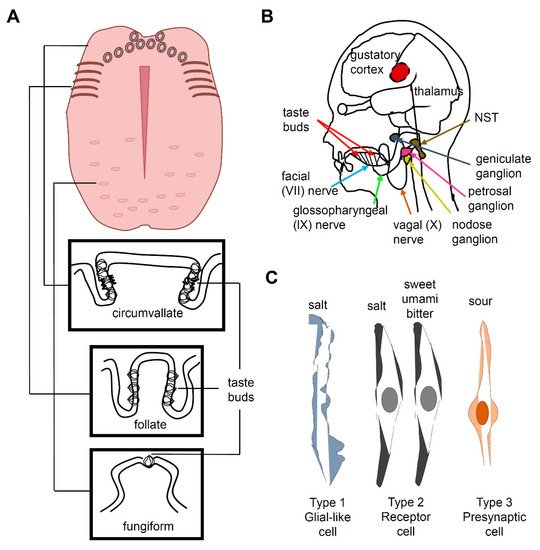
2. Salt Taste in Mammals
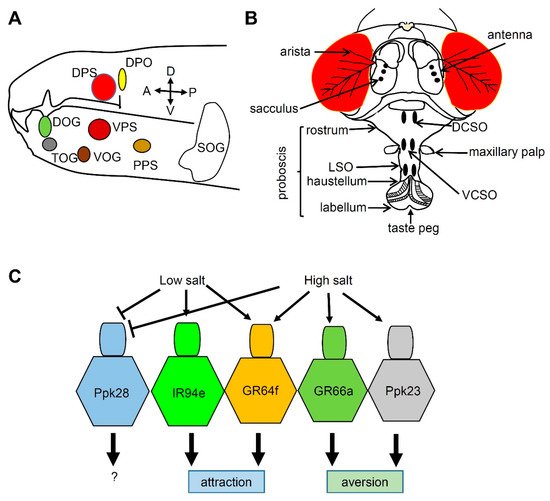
3. Salt Taste in Drosophila
This entry is adapted from the peer-reviewed paper 10.3390/metabo11030175
References
- Matthews, R.W. The Insects: Structure and Function; Oxford University Press: Oxford, UK, 1971.
- Joseph, R.M.; Carlson, J.R. Drosophila Chemoreceptors: A Molecular Interface between the Chemical World and the Brain. Trends Genet. 2015, 31, 683–695.
- Chandrashekar, J.; Kuhn, C.; Oka, Y.; Yarmolinsky, D.A.; Hummler, E.; Ryba, N.J.; Zuker, C.S. The cells and peripheral representation of sodium taste in mice. Nature 2010, 464, 297–301.
- Lindemann, B. Receptors and transduction in taste. Nature 2001, 413, 219–225.
- Oka, Y.; Butnaru, M.; von Buchholtz, L.; Ryba, N.J.; Zuker, C.S. High salt recruits aversive taste pathways. Nature 2013, 494, 472–475.
- Kaushik, S.; Kumar, R.; Kain, P. Salt an Essential Nutrient: Advances in Understanding Salt Taste Detection Using Drosophila as a Model System. J. Exp. Neurosci. 2018, 12, 1179069518806894.
- Chandrashekar, J.; Hoon, M.A.; Ryba, N.J.; Zuker, C.S. The receptors and cells for mammalian taste. Nature 2006, 444, 288–294.
- Nomura, K.; Nakanishi, M.; Ishidate, F.; Iwata, K.; Taruno, A. All-electrical Ca2+-independent signal transduction mediates attractive sodium taste in taste buds. Neuron 2020, 106, 816–829.
- Ohmoto, M.; Jyotaki, M.; Foskett, J.K.; Matsumoto, I. Sodium–Taste Cells Require Skn-1a for Generation and Share Molecular Features with Sweet, Umami, and Bitter Taste Cells. Eneuro 2020, 7.
- Zhao, G.Q.; Zhang, Y.; Hoon, M.A.; Chandrashekar, J.; Erlenbach, I.; Ryba, N.J.; Zuker, C.S. The receptors for mammalian sweet and umami taste. Cell 2003, 115, 255–266.
- Xu, H.; Staszewski, L.; Tang, H.; Adler, E.; Zoller, M.; Li, X. Different functional roles of T1R subunits in the heteromeric taste receptors. Proc. Natl. Acad. Sci. USA 2004, 101, 14258–14263.
- Nelson, G.; Chandrashekar, J.; Hoon, M.A.; Feng, L.; Zhao, G.; Ryba, N.J.; Zuker, C.S. An amino-acid taste receptor. Nature 2002, 416, 199–202.
- Vegezzi, G.; Anselmi, L.; Huynh, J.; Barocelli, E.; Rozengurt, E.; Raybould, H.; Sternini, C. Diet-induced regulation of bitter taste receptor subtypes in the mouse gastrointestinal tract. PLoS ONE 2014, 9, e107732.
- Toda, Y.; Nakagita, T.; Hayakawa, T.; Okada, S.; Narukawa, M.; Imai, H.; Ishimaru, Y.; Misaka, T. Two distinct determinants of ligand specificity in T1R1/T1R3 (the umami taste receptor). J. Biol. Chem. 2013, 288, 36863–36877.
- Avau, B.; Rotondo, A.; Thijs, T.; Andrews, C.N.; Janssen, P.; Tack, J.; Depoortere, I. Targeting extra-oral bitter taste receptors modulates gastrointestinal motility with effects on satiation. Sci. Rep. 2015, 5, 1–12.
- Liman, E.R.; Zhang, Y.V.; Montell, C. Peripheral Coding of Taste. Neuron 2014, 81, 984–1000.
- Roper, S.D.; Chaudhari, N. Taste buds: Cells, signals and synapses. Nat. Rev. Neurosci. 2017, 18, 485–497.
- Hill, D.L.; Mistretta, C.M.; Bradley, R.M. Developmental changes in taste response characteristics of rat single chorda tympani fibers. J. Neurosci. 1982, 2, 782–790.
- LopezJimenez, N.D.; Cavenagh, M.M.; Sainz, E.; Cruz-Ithier, M.A.; Battey, J.F.; Sullivan, S.L. Two members of the TRPP family of ion channels, Pkd1l3 and Pkd2l1, are co-expressed in a subset of taste receptor cells. J. Neurochem. 2006, 98, 68–77.
- Kataoka, S.; Yang, R.; Ishimaru, Y.; Matsunami, H.; Sévigny, J.; Kinnamon, J.C.; Finger, T.E. The candidate sour taste receptor, PKD2L1, is expressed by type III taste cells in the mouse. Chem. Senses 2008, 33, 243–254.
- Lossow, K.; Hermans-Borgmeyer, I.; Behrens, M.; Meyerhof, W. Genetic labeling of Car4-expressing cells reveals subpopulations of type III taste cells. Chem. Senses 2017, 42, 747–758.
- AlJulaih, G.H.; Lasrado, S. Anatomy, Head and Neck, Tongue Taste Buds; Statpearls: Treasure Island, FL, USA, 2019.
- Breslin, P.A.; Spector, A.C. Mammalian taste perception. Curr. Biol. 2008, 18, R148–R155.
- Adler, E.; Hoon, M.A.; Mueller, K.L.; Chandrashekar, J.; Ryba, N.J.; Zuker, C.S. A novel family of mammalian taste receptors. Cell 2000, 100, 693–702.
- Corson, S.L.; Hill, D.L. Chorda Tympani Nerve Terminal Field Maturation and Maintenance Is Severely Altered Following Changes to Gustatory Nerve Input to the Nucleus of the Solitary Tract. J. Neurosci. 2011, 31, 7591–7603.
- Bachmanov, A.A.; Beauchamp, G.K.; Tordoff, M.G. Voluntary consumption of NaCl, KCl, CaCl 2, and NH 4 Cl solutions by 28 mouse strains. Behav. Genet. 2002, 32, 445–457.
- Brand, J.G.; Teeter, J.H.; Silver, W.L. Inhibition by amiloride of chorda tympani responses evoked by monovalent salts. Brain Res. 1985, 334, 207–214.
- Heck, G.L.; Mierson, S.; DeSimone, J.A. Salt taste transduction occurs through an amiloride-sensitive sodium transport pathway. Science 1984, 223, 403–405.
- Canessa, C.M.; Schild, L.; Buell, G.; Thorens, B.; Gautschi, I.; Horisberger, J.-D.; Rossier, B.C. Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits. Nature 1994, 367, 463–467.
- Lewandowski, B.C.; Sukumaran, S.K.; Margolskee, R.F.; Bachmanov, A.A. Amiloride-Insensitive Salt Taste Is Mediated by Two Populations of Type III Taste Cells with Distinct Transduction Mechanisms. J. Neurosci. 2016, 36, 1942–1953.
- Grant, G.J.; Coca, C.; Zhao, X.-M.; Helms, M.N. Oxidized Glutathione Increases Delta-Subunit Expressing Epithelial Sodium Channel Activity in Xenopus laevis Oocytes. Emed. Res. 2020, 2, 100008.
- Wichmann, L.; Vowinkel, K.S.; Perniss, A.; Manzini, I.; Althaus, M. Incorporation of the δ-subunit into the epithelial sodium channel (ENaC) generates protease-resistant ENaCs in Xenopus laevis. J. Biol. Chem. 2018, 293, 6647–6658.
- Baldin, J.-P.; Barth, D.; Fronius, M. Epithelial na+ channel (ENaC) Formed by one or two subunits forms functional channels that respond to shear force. Front. Physiol. 2020, 11, 141.
- Pitzer, A.L.; Van Beusecum, J.P.; Kleyman, T.R.; Kirabo, A. ENaC in Salt-Sensitive Hypertension: Kidney and Beyond. Curr. Hypertens. Rep. 2020, 22, 1–10.
- Lu, B.; Breza, J.M.; Contreras, R.J. Temperature Influences Chorda Tympani Nerve Responses to Sweet, Salty, Sour, Umami, and Bitter stimuli in mice. Chem. Senses 2016, 41, 727–736.
- Roebber, J.K.; Roper, S.D.; Chaudhari, N. The Role of the Anion in Salt (NaCl) Detection by Mouse Taste Buds. J. Neurosci. 2019, 39, 6224–6232.
- Ninomiya, Y. Reinnervation of cross-regenerated gustatory nerve fibers into amiloride-sensitive and amiloride-insensitive taste receptor cells. Proc. Natl. Acad. Sci. USA 1998, 95, 5347–5350.
- Stocker, R.F. The organization of the chemosensory system in Drosophila melanogaster: A rewiew. Cell Tissue Res. 1994, 275, 3–26.
- Lee, Y.; Poudel, S. Taste sensation in Drosophila melanogaster. Hanyang Med. Rev. 2014, 34, 130–136.
- Rimal, S.; Lee, Y. The multidimensional ionotropic receptors of Drosophila melanogaster. Insect Mol. Biol. 2018, 27, 1–7.
- Zhang, Y.V.; Ni, J.; Montell, C. The Molecular Basis for Attractive Salt-Taste Coding in Drosophila. Science 2013, 340, 1334–1338.
- Zelle, K.M.; Lu, B.; Pyfrom, S.C.; Ben-Shahar, Y. The Genetic Architecture of Degenerin/Epithelial Sodium Channels in Drosophila. G3 Genes Genomes Genet. 2013, 3, 441–450.
- Jaeger, A.H.; Stanley, M.; Weiss, Z.F.; Musso, P.-Y.; Chan, R.C.; Zhang, H.; Feldman-Kiss, D.; Gordon, M.D. A complex peripheral code for salt taste in Drosophila. Elife 2018, 7, e37167.
- Liu, L.; Johnson, W.A.; Welsh, M.J. Drosophila DEG/ENaC pickpocket genes are expressed in the tracheal system, where they may be involved in liquid clearance. Proc. Natl. Acad. Sci. USA 2003, 100, 2128–2133.
- Lee, M.J.; Sung, H.Y.; Jo, H.; Kim, H.-W.; Choi, M.S.; Kwon, J.Y.; Kang, K. Ionotropic Receptor 76b Is Required for Gustatory Aversion to excessive Na+ in Drosophila. Mol. Cells 2017, 40, 787.
- Lee, Y.; Poudel, S.; Kim, Y.; Thakur, D.; Montell, C. Calcium Taste Avoidance in Drosophila. Neuron 2018, 97, 67–74.e64.
- Jiao, Y.; Moon, S.J.; Wang, X.; Ren, Q.; Montell, C. Gr64f Is Required in Combination with Other Gustatory Receptors for Sugar Detection in Drosophila. Curr. Biol. 2008, 18, 1797–1801.
- Moon, S.J.; Köttgen, M.; Jiao, Y.; Xu, H.; Montell, C. A Taste Receptor Required for the Caffeine Response In Vivo. Curr. Biol. 2006, 16, 1812–1817.
- Cameron, P.; Hiroi, M.; Ngai, J.; Scott, K. The molecular basis for water taste in Drosophila. Nature 2010, 465, 91–95.
- Kim, H.; Jeong, Y.T.; Choi, M.S.; Choi, J.; Moon, S.J.; Kwon, J.Y. Involvement of a Gr2a-Expressing Drosophila Pharyngeal Gustatory Receptor Neuron in Regulation of Aversion to High-Salt Foods. Mol. Cells 2017, 40, 331–338.
