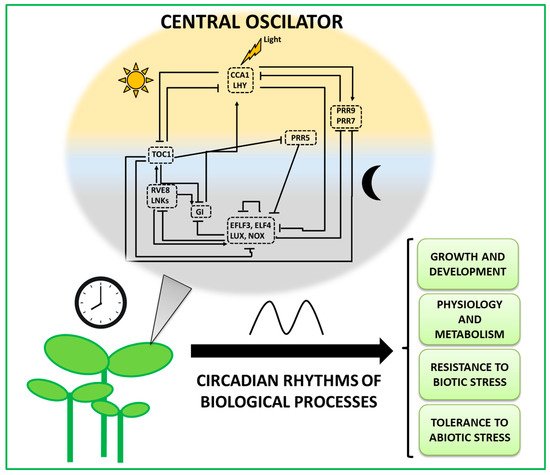
During the last few decades, much effort has been put into studying whether information of the Arabidopsis circadian clock can be extended to elucidate the molecular elements of the circadian core clock in
Brassica crop species. The first studies began by identifying the natural allelic variation in clock function based on quantitative trait locus (QTL) analysis [
8,
31]. These studies investigated cotyledon movement from
B. oleracea and
B. rapa to detect genetic loci affecting the circadian period. Comparative genomic analysis based on collinearity between
Brassica and Arabidopsis also allowed the identification of candidate genes known to regulate the period in Arabidopsis that may account for the additive circadian effects of specific QTL [
32,
33,
34]. This includes gene families encoding
PRRs,
TOC1,
CCA1, and
LHY.
Brassica species experienced an extra whole-genome triplication event compared with Arabidopsis. Therefore, as was expected, genes contributing to circadian clock function were retained in multiple copies in
Bassica species. Song et al. [
35] cloned and analyzed
CCA1 genes from seven inbred lines and one cultivar of cabbage (
Brassica oleracea). Two types of
CCA1 alleles were detected and related to freezing-tolerant cabbage traits. In another study, the natural variation in
CCA1 was associated with the flowering time in
B. rapa and a high level of sequence variation was identified [
36]. Genetic mapping and analysis of families of heterogeneous inbred lines showed that the natural variation in
GI is responsible for a major quantitative trait locus in the circadian period in
B. rapa. Loss-of-function mutations of
GI from
B. rapa and
B. oleracea cultivars confer delayed flowering, perturbed circadian rhythms in leaf movement, caused leaf senescence, and increased freezing and salt tolerance, consistent with the effects of similar mutations in Arabidopsis [
37,
38].
More recently, Greenham et al. [
6] and Kim et al. [
39] performed high-resolution circadian transcriptome experiments to elucidate the
B. rapa circadian network. They found that genes related to the clock displayed distinct phases, increasing or decreasing in regular patterns. In addition, the different copies of duplicated and triplicated genes did not necessarily all behave in the same way. Many of the copies had different rhythms, and some increased and decreased in patterns totally opposite to their counterparts. Not only did the daily patterns differ, but responses to stressors were also altered. Comparing these patterns to the patterns seen in Arabidopsis revealed that often, one
B. rapa gene behaved just like its Arabidopsis equivalent, while its copies had evolved new behaviors. The authors conclude that different behaviors of the copies of each gene in
B. rapa, relative to its biological clock, allow this plant to grow in different environments with varying temperatures and day lengths.
The clock-regulated genes identified in
Brassica are required for primary and secondary metabolism, photosynthesis, cold stress, and response to biotic stimulus [
6,
34,
40,
41], which strongly evidences the role of the circadian clock in vegetative growth and plant physiological processes. In line with that, recent works are starting to show that the circadian core clock genes in
Brassica crops underlie QTLs that have been related to beneficial influences on key agricultural traits, especially flowering time but also yield, biomass, and growth [
42,
43,
44,
45]. In addition, new insights into the circadian clock regulation of other fundamental plant processes, such as responses to abiotic and biotic stresses, could help to guide future work in targeting genes to improve crop growth and stress resilience.