Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Pharmacology & Pharmacy
Clopidogrel is a widely-used antiplatelet drug. It is important for the treatment and prevention of coronary heart disease. Clopidogrel can effectively reduce platelet activity and therefore reduce stent thrombosis. However, some patients still have ischemic events despite taking the clopidogrel due to the alteration in clopidogrel metabolism attributable to various genetic and non-genetic factors. This review aims to summarise the mechanisms and causes of clopidogrel resistance (CR) and potential strategies to overcome it.
- clopidogrel 2
- antiplatelet 3
- clopidogrel resistance 4
- CYP2C19 polymorphism 5
- personalized medicine
1. Definition of Clopidogrel Resistance
There is currently no uniform definition of CR, but the most accepted is that the drug has lost its target of the action. It is generally believed that CR means that a patient still has a thrombotic event after receiving clopidogrel treatment, and laboratory tests show that platelet function is not inhibited [31]. Some researchers refer to it as clinical resistance among patients who have experienced thromboembolism and other adverse events following long-term oral clopidogrel therapy [1]. The incidence of CR varies among different regions and races. According to literature reports, the incidence of CR in Western countries is 5 to 44%, while in Asian populations, it may be as high as 20 to 65% [1,32].
There are several methods commonly used to evaluate platelet function. The oldest and more accurate way is optical turbidimetry, which is often considered the gold standard. This method assesses the responsiveness of platelets to ADP through the function of P2Y1 and P2Y12 receptors. However, because of the repetition rate and the lack of a specific P2Y12 pathway, its use is limited. At present, vasodilator-stimulated phosphoprotein (VASP) phosphorylation assay (VerifyNow) and bedside monitoring are widely used due to the relatively easy operation [11,33]. Tantry and colleagues (2014), in their follow-up studies on CR, confirmed that the available evidence does not support routine screening for hypo/non-responsiveness in patients who started treatment with clopidogrel [34]. So far, there is still a lack of standard experimental methods for diagnosing CR. Clinically, platelet function can be tested to determine the patient’s platelet reaction after medication intake to identify the potential risk of increased cardiovascular or bleeding events. The incidence of CR in elderly patients may be higher than that in younger patients, and the risk of bleeding with clopidogrel is also increased [35,36,37].
The use of platelet function tests (PFTs) to allocate a better selection of antiplatelet drugs to patients with cardiovascular disease has been discussed over the past ten years [38]. These studies mitigated the escalation of antiplatelet therapy according to the results of PFTs for potential clinical benefit. Furthermore, the 2011 American College of Cardiology/American Heart Association guidelines issued a Class IIb recommendation for the use of PFTs among patients taking P2Y12 inhibitors [39]. Still, this classification was downgraded to a Category III recommendation in 2016 [40]. In ACS cases, the latest European guidelines indicate that de-escalation, but not escalation, of P2Y12 inhibitors directed by PFT, with a Class IIb rating, can be considered [41].
2. Factors Associated with CR
The mechanism of CR is still unclear. Relevant studies have shown that CR may be influenced by various factors such as race, age, weight, genetic polymorphism, drug interaction, diabetes, inflammation, immature platelets, atherosclerosis, medication compliance and other factors. Despite these various contributing factors of clopidogrel resistance, the exact mechanism is currently unknown [1,37,42,43,44,45,46,47,48,49].
2.1. Gene Polymorphism
Many studies have been done to determine the relationship between P2Y12 receptor gene polymorphism and CR (Table 2). Zoheir et al. (2013) found that P2Y12 receptor gene polymorphism is closely related to platelet activity [50]. The P-gp encoded by ABCB1 regulates the absorption of clopidogrel in the intestines. Earlier studies by Mega et al. (2009) found that ABCB1 gene polymorphism affects the degree of platelet inhibition, which is closely related to the risk of major adverse cardiac events (MACE) [5]. However, in recent years, studies on the Chinese population have shown no association between ABCB1 gene polymorphism and CR [51,52,53].
Table 2. Genetic polymorphism distribution and allele frequencies in clopidogrel-resistant and non-clopidogrel-resistant groups.
CYP3A4/5 are among the essential enzymes in clopidogrel activation. Previously, Lau et al. (2004) have shown that lower CYP3A4 activity, determined using an erythromycin breath test, is associated with a lower antiplatelet effect of the drug [54].
One study aimed to determine the effect of the CYP3A homologs of sub-enzymes (allelic variants of CYP3A4 * 22 and CYP3A5 * 3) on the efficacy of clopidogrel in patients with ACS undergoing percutaneous coronary intervention. The study results found that CYP3A4 / 5 activity was not associated with platelet aggregation rates, as well as the genotyping and phenotyping of CYP3A4 / CYP3A5 did not predict the antiplatelet effect of clopidogrel. The researcher recommended more extensive research to prove its clinical relevance [55].
The genetic variations in CYP450 isoenzymes genes (CYP1A2, CYP2B6, CYP2C9, CYP2C19, and CYP3A4), which are involved in drug metabolism, can influence the variation of pharmacodynamic response to clopidogrel, especially the genetic variation in the CYP2C19 isoenzyme. This enzyme contributes significantly to the two sequential oxidative steps in the biotransformation of clopidogrel into active metabolites [56,57]. Hence, genetic polymorphism of CYP2C19 could play a crucial role in wide inter-individual and inter-ethnic variabilities in clinical response towards clopidogrel [58,59,60,61].
The choice of antiplatelet therapy (clopidogrel, ticagrelor, or prasugrel) based on individual patient characteristics, such as treatment choice based on genetic data related to clopidogrel metabolism as well as considerations regarding the clinical features of patients may result in a significantly lower rate of ischemic and hemorrhagic events compared to usual practice [70]. The choice of antiplatelet therapy based on both CYP2C19 gain of function (GOF) and loss of function (LOF) alleles appears to be a preferred approach over universal clopidogrel and universal variant P2Y12 inhibitor therapy for ACS patients with PCI [71,72]. CYP2C19-guided escalation and de-escalation are common as clopidogrel persistence in nonfunctional allele carriers is associated with adverse outcomes [73].
Genetic polymorphisms in CYP2C19 were classified into groups and referred to as alleles. The preliminarily identified alleles include 36 alleles such as CYP2C19 *1,*2, *3, *4, *5, *6, *7 or *8 etc. of which the most significant impact on clopidogrel is *2/*3 mutation sites (weak metabolites) and *17 mutation sites (strong metabolites). The frequency of other variations in most population groups is low [74]. According to clinical guidelines issued by the Clinical Pharmacogenetics Implementation Consortium (CPIC), genotype-related individual variability in metabolic enzyme function is divided into four predicted CYP2C19 metabolic phenotypes: Poor metabolisers (PMs), intermediate metabolisers (IMs), Extensive metabolisers (EMs), and Ultrarapid metabolisers (UMs) [75] (Table 3).
Table 3. The categorisation of the predicted CYP2C19 metabolic phenotypes based on the CYP2C19 genotypes [75].
Many studies have reported wide inter-ethnic variability in CYP2C19 polymorphism. Asian populations (~ 55.0 to 70.0%) have a higher prevalence rate of CYP2C19 LOF variant alleles (CYP2C19 *2 and *3) as compared with white populations (~ 25.0 to 35.0%) and black populations (~35.0 to 45.0%) [76,77]. On the other hand, Asian populations (~4.0%) have a low prevalence of the CYP2C19 GOF variant allele (CYP2C19 *17) as compared to white populations (~18.0%) [78,79].
Recent studies have reported a variation in the prevalence of individuals carrying CYP2C19 alleles among the Asian population (Table 4). The CYP2C19 * 2 allele was found in individuals of the selected countries, with prevalence rates ranging between 4.0–59.6%, with an average prevalence rate of 23.00%. The percentage prevalence of CYP2C19 * 2 allele in Saudi Arabia, Qatar and Jordan was less than 10% (residents of the Arabian Peninsula), which is low compared to others. Meanwhile, the CYP2C19 * 3 allele prevalence was found at rates up to 0–13.03% with an average prevalence rate of 4.61%. It is noticed that the spread of this allele is higher in the countries of Southeast and East Asia. Still, its prevalence rates are lower in India, located in the south of Asia, Russia, which is in its north and most countries in West Asia, excluding Turkey. From the CYP2C19 * 17 allele prevalence data, it is noticed that the prevalence rates ranged between (1- 28.72) %, with an average rate of 15.18%, as it is seen here that there are high prevalence rates in the North, South and West Asia. Medium to low rates are observed in some Central and Southeast Asia (Figure 2).
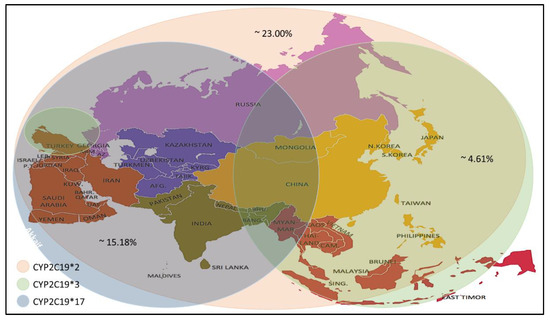
Figure 2. Prevalence of the CYP2C19 * 2/*3/*17 alleles in the Asian population.
Table 4. CYP2C19 allele frequencies (* 2, * 3 and * 17) % among Asian ethnic groups.
In general, the high allele frequency of CYP2C19 * 2 and * 17 in the Asian population led to the recommendation of a pre-treatment test to monitor for clopidogrel response, dose and to avoid adverse drug reactions after treatment.
2.2. Drug Interactions
It is known that clopidogrel is converted into an effective product through the metabolic pathway mediated by CYP enzymes. This process involves a variety of isoenzymes. Such as CYP2C19, CYP3A4, CYP1A2, CYP2C9, etc., but the most important ones are CYP3A4 (~40%) and CYP2C19 (~45%) that contribute to the formation of the active metabolite of clopidogrel; so, the combined use of CYP3A4 and CYP2C19 inhibitors may affect the metabolism of clopidogrel [57,97]. Besides clopidogrel, the CYP3A4 pathway also metabolises statins and calcium channel blockers, and the CYP2C19 pathway metabolises proton pump inhibitors (PPIs) [28,98]. Figure 3 illustrates the mechanism by which these three compounds affect clopidogrel.
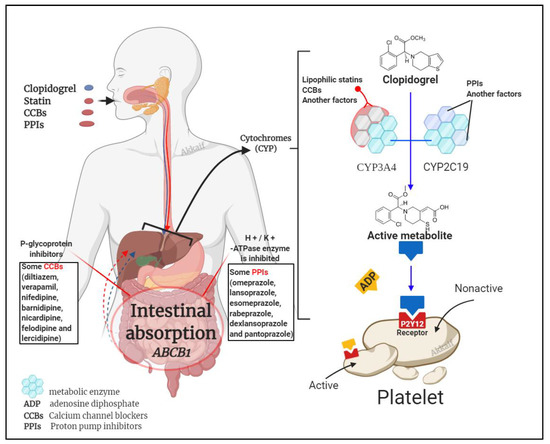
Figure 3. Drug interaction mechanism of clopidogrel with statins, calcium channel blockers (CCBs) and proton pump inhibitors (PPIs).
2.3. Dose Factors
The anti-platelet effect of clopidogrel is dose-dependent [17]. The 300 mg loading dose of clopidogrel reaches a steady state after 4 to 24 h. If there is no load, it takes 4 to 7 days to reach a steady-state [118]. Allier et al. found that the antiplatelet effect of clopidogrel 600 mg administered for the first time was equivalent to that of long-term 75 mg patients. Clopidogrel 600 mg administered during long-term treatment can further inhibit platelet aggregation [119]. Due to the increase in thrombus load before treatment, the standard loading dose is not enough to achieve effective platelet inhibition for patients with severe symptoms. Therefore, CR will still occur with conventional-dose treatment [120].
2.4. Other Factors
Among other factors, patients’ compliance also directly affects the effectiveness of clopidogrel. Other than that, the antiplatelet effect of clopidogrel is limited in type 2 diabetes patients because this disease is often associated with atherosclerotic disease manifestations; clopidogrel is commonly used in these patients [121]. Diabetes is also a risk factor for reduced antiplatelet effects by clopidogrel [121,122]. There is also a vital relationship found between the level of inflammatory factors and CR caused by abnormal platelet function [123,124,125].
3. Strategies to Overcome CR
3.1. Increase the Dose of Clopidogrel
Increasing the dose can increase the biological effect of clopidogrel and reduce the incidence of CR. Simultaneously, large doses of clopidogrel can reduce patients’ platelet aggregation rate with CR [126]. For PCI patients, the 600 mg loading dose has a faster response than the 300 mg loading dose and has a more substantial platelet inhibitory effect. In this way, the incidence of CR is significantly reduced [127,128]. At the same time, studies have shown that CR or platelet hyperresponsiveness is still common after the administration of clopidogrel 600 mg load, but increasing the dose can reduce the risk of death from cardiovascular disease, myocardial infarction, and stent thrombosis [5]. In patients with stable coronary heart disease, CYP2C19*2 heterozygous carriers taking 225 mg of clopidogrel per day were shown to achieve the same antiplatelet effect with CYP2C19 wild-type patients taking 75 mg of clopidogrel per day. In contrast, CYP2C19*2 homozygous patients cannot achieve the desired antiplatelet effect even if they take the 300 mg clopidogrel maintenance dose [129]. CR in patients treated with PCI between high maintenance dose (150 mg · d −1) than conventional maintenance dose (75 mg · d −1) can more effectively prevent major adverse cardiac events (MACE). In the 1-month follow-up after PCI, the incidence of in-stent thrombosis was lower among the group receiving 150 mg · d −1 as compared to the group receiving 75 mg · d −1 (1.1% and 4.9%, p = 0.03). Simultaneously, cardiovascular events incidence was also significantly lower in the group with higher doses (2.7% and 7.6%, p = 0.03) [130]. However, some studies have shown that high-dose clopidogrel after PCI did not reduce the mortality of cardiovascular events or stent thrombosis incidence than standard doses [131]. Moreover, high-dose clopidogrel may lead to an increased probability of bleeding complications; therefore, the use of high-dose clopidogrel maintenance treatment to avoid treatment resistance requires further research.
3.2. Combined Use of Other Antiplatelet Drugs
Ainetdinova et al. [132] found that the probability of resistance to aspirin, clopidogrel, and the combination of these two drugs were 25.7%, 17.1%, and 5.7%, respectively. Therefore, DAPT with aspirin and clopidogrel was shown to reduce the occurrence of drug resistance. Another potential combination therapy uses the GPIIb/IIIa receptor antagonists (such as abciximab, tirofiban and eptifibatide), which can directly block the final pathway of platelet activation, adhesion, and aggregation. Based on clopidogrel therapy, the combined use of GPIIb/IIIa receptor antagonists can further inhibit platelet aggregation [133,134].
3.3. Replacement of New P2Y12 Receptor Antagonists
The new P2Y12 inhibitors, ticagrelor and prasugrel, will substantially reduce platelet hyperresponsiveness and improve clinical outcomes relative to the regular clopidogrel dose. Most patients who do not respond to clopidogrel can significantly inhibit the platelet aggregation rate after switching to prasugrel [135] because prasugrel can better inhibit ADP-induced platelet aggregation, which is faster and stronger than clopidogrel. The longer-lasting antiplatelet effect of prasugrel can significantly reduce the occurrence of ischemic events [136]. On the other hand, ticagrelor does not require liver metabolism and not affected by CYP2C19 gene polymorphism. It was also shown to significantly reduce mortality related to cardiovascular events, myocardial infarction [137]. A study showed that in STEMI patients undergoing PCI for the first time, a loading dose of 180 mg of ticagrelor was more effective than a loading dose of 600 mg of clopidogrel in reducing microvascular damage [138]. There is also literature mentioning that cangrelor has a powerful platelet inhibitory effect. Its effect may be more significant than clopidogrel. Moreover, its half-life is shorter, does not require liver activation, and is a direct antagonist of P2Y12 [26].
3.4. Other Management of CR
Active control of blood sugar in patients with coronary heart disease can reduce the incidence of CR. Avoiding the simultaneous application of other drugs that require CYP metabolisms, such as statins, calcium channel blockers, and PPI, would ensure a better response to clopidogrel therapy.
In a randomised trial of TROPICAL-ACS [139,140], a targeted de-escalation regimen with early switching from prasugrel to clopidogrel was established as an effective alternative treatment strategy in ACS patients. However, the study found that patient age was the primary determinant of outcome after PCI, [141,142], especially when using P2Y12 receptor inhibitors during and after PCI [143,144]. Therefore, TROPICAL-ACS performed a randomised assessment of the effect of age on reducing the escalation of antiplatelet therapy. Significant variation was found among the younger patients who showed an increased net clinical benefit resulting from reduced bleeding complications. These results suggest that targeted de-escalation may be a safe and attractive alternative therapy concept for all ACS patients after PCI, while a significant bleeding benefit could be achieved in younger patients [145].
This entry is adapted from the peer-reviewed paper 10.3390/molecules26071987
This entry is offline, you can click here to edit this entry!
