Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) proteins is a natural adaptive immune system that some bacterial and most archaeal species present to defend themselves against invading bacteriophages, which works on the basis of sequence complementarity via cleavage. Genetic engineering using CRISPR/Cas systems enables accurate and precise genomic modifications. The easiness and rapidity of execution, low cost, reproducibility and efficiency turns understandable why it is the system of choice for many genome engineering applications in several fields using different organisms.
- CRISPR/Cas systems
- viral vectors
- gene editing
- plant genome engineering
- viral resistance
1. CRISPR: From a Natural Bacterial Immune System to a Gene Editing Tool
CRISPR systems may be divided into two main classes (I and II) and six different types (I to VI), defined by the nature of the nucleases complex and the mechanism of targeting, each presenting a unique nuclease Cas protein. Class I systems are multicomponent systems composed of multiple effectors; these systems are subdivided into types I, III, and IV. Class II systems include the types II, V, and VI and are single-component systems consisting of a single effector guided by the CRISPR RNA (crRNA) [1].
The CRISPR/Cas9, belonging to class II, is based on the immune system of Streptococcus pyogenes. It consists of the capacity of the bacteria to acquire pieces of DNA from an invading phage or plasmid and incorporating them in their own DNA, which will further serve to guide Cas9 to cleave homologous RNA, leading to immediate RNA disruption and further specific RNA disruption in subsequent invasions, thus providing immunity to the bacterial cell [2]. The mechanism involved in this natural immune system is very simple and has been the basis for the most developed CRISPR/Cas genome-editing platform.
The first steps of CRISPR/Cas9 as a successful editing tool, started with the possibility of engineering into a single RNA chimera (sgRNA), two noncoding RNAs essential for CRISPR, crRNA, and trans-activating crRNA (tracrRNA) [3]. crRNA is the genomic complementary region, i.e., the target for Cas (the programmable portion defined by the user) and tracrRNA is the RNA sequence that provides the stem loop structure to bound Cas. This has simplified gene editing using CRISPR/Cas9, which can now be accomplished by introducing two components in the same cell: the sgRNA and the Cas protein [3] and led to efficient genetic manipulation in a wide array of plants, becoming the most promising, versatile, and powerful tool for plant improvement [4].
In CRISPR/Cas9 system (Figure 1), first Cas9 binds to the sgRNA to create the Cas9-sgRNA duplex which becomes catalytically active and directs the RNA-guided DNA endonuclease Cas9 to target. For target recognition and cleavage, it is also required the presence of a Protospacer Adjacent Motif (PAM) positioned 3–4 nucleotides downstream of the 3′ end of the target sequence, which differs depending on the species of Cas9 (this sequence consists of NGG in S. pyogenes) [3][5]. Once the PAM sequence is recognized by the Cas9-sgRNA complex, and the crRNA portion within the sgRNA (the 5′ most 20 nts) anneals to the genomic DNA through Watson–Crick base pairing, it will cleave both DNA strands, three bases upstream of the PAM, creating sequence-specific blunt end double-stranded breaks (DSBs) at target site. When a DSB in the DNA is created, the host cell repairs it via evolutionary conserved DNA pathways such as error-prone non-homologous end-joining (NHEJ) and homology-directed repair (HDR).
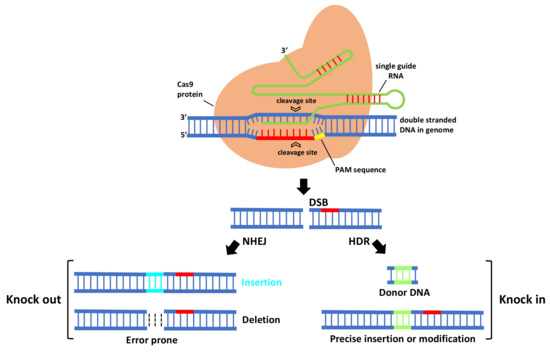
Figure 1. The mechanism of CRISPR-Cas9-mediated genome engineering in plants. A single guide RNA recognizes a region in the genome followed by a PAM sequence, and recruits a Cas9 protein that will cleave DNA, creating a double-stranded break that is repaired by error-prone non-homologous end-joining (NHEJ) and homology-directed repair (HDR).
NHEJ creates insertions or deletions (indels) at the target site that, if within the protein coding region, can cause a frameshift mutation that eliminates gene expression, leading to gene knock out [6]. HDR is a more precise method for DSB repair; it requires, besides sgRNA and Cas, a donor repair template with ends homologous to each border of the target site sequence. When a repair template is provided, HDR will result in the introduction of new sequences at breaking site and a knock in occurs [6]. For producing specific desired mutations and genomic replacement, DSBs should be repaired by HDR pathway. More recently, a new generation of CRISPR is being developed by fusing nuclease DNA targeting proteins with deactivated nuclease domains, with enzymes to enable direct conversion of a single DNA nucleotide into another [7] without the need of DSB formation.
Genetic engineering using CRISPR/Cas systems enables accurate and precise genomic modifications. Moreover, this strategy can be used to target different sequences simultaneously with high efficiency [8], achieving a broader result, as for example immunity against different pathogens.
The easiness and rapidity of execution, low cost, reproducibility and efficiency turns understandable why it is the system of choice for many genome engineering applications in several fields using different organisms. The possibility of using Cas proteins with deactivated nuclease domains can contribute to a broader application of CRISPR such as regulating gene transcription and inducing targeted epigenic modifications [9]. In addition, CRISPR has shown to have potential for other applications besides genome engineering, such as studies on gene functions and diagnostics. CRISPR/LwaCas13a system was able to highly select and detect up to a single copy of RNA [10], which may be a very interesting starting point to develop a far more sensitive method than currently available methods, for the detection of RNA viruses, including qPCR [11].
In plants, this technology has been used for plant breeding including nutrition enhancement and plant resistance against several agents such as fungi, bacteria, and viruses in many crop plants—including rice [12], tomato [13], citrus [14][15], wheat [16], and maize [17][18]—proving its potential to transform agriculture and enhancing world food safety.
2. CRISPR to Engineer Plant Virus Resistance
Due to the devastating losses that plant viruses cause, it is not surprising that CRISPR/Cas technologies have been applied to develop plant resistance against viral pathogens.
Plant viral resistance using CRISPR/Cas systems can been achieved either through manipulation of plant genome (plant-mediated resistance), or virus genome (virus-mediated resistance).
The CRISPR/Cas technology was initially thought to be exclusively applied to DNA, which, in terms of its use for plant viral resistance through manipulation of viral genome, would be restricted to DNA viruses. However, thanks to the discovery of RNA-targeting CRISPR/Cas effectors that efficiently target and cleave single-stranded RNAs, an exciting opportunity has been opened for achieving plant resistance also against RNA viruses, which are most of the plant viruses known [19][20].
Below we present several studies that report the use of the CRISPR/Cas system to engineer plant resistance against several viruses, either by acting on plant genome (plant mediated resistance) or on viral genomes (virus mediated resistance). These studies have shown the capacity of CRISPR to confer efficient and durable molecular immunity to plants against viruses that rely on the integrity of their genome at some point of their replication cycle [21][22][23][24][25].
2.1. CRISPR for Plant Mediated Resistance
Plant viruses are dependent on the host’s machinery for their replication, since they interact with many host factors required for viral replication and movement inside plants, essential to complete their cycle of infection [26]. CRISPR/Cas allows the mutation/deletion of recessive genes that encode critical host factors for viral infection, conferring recessive resistance, which, as an inherited characteristic is very durable [27].
Considerable knowledge has been generated on the genetics of plant disease resistance and many plant genes have been discovered as essential for viral infections and have been the focus for the development of plant resistance using transgenic approaches [28][29][30]. These studies have provided many valuable potential targets for genome editing and genes—such as the translation initiation-like factors elF4E, elF4G, and their isoforms—that have shown to be directly involved in the infection process of viruses. Those genes are being subjected to targeted mutations introduced by CRISPR to engineer plant resistance [31]. In fact, any host gene encoding a factor required by the virus is a potential target for CRISPR.
This approach is interesting as it allows that Cas9, as well as other endonucleases which target DNA, to be used to provide plant resistance to RNA viruses by mutating host factors/genes associated to viral pathogenesis in the plant [32]. In addition, CRISPR for plant mediated resistance does not require the maintenance of a transgene for Cas9 and sgRNA in the plant genome, engineering transgenic-free virus-resistant plants [33][24][32].
Several studies have achieved plant mediated resistance against viruses using CRISPR/Cas9 (Table 1). For example, specific mutations were introduced in Arabidopsis thaliana, causing the knock out of elF(iso)4E gene, which resulted in a stable resistance against Turnip mosaic virus (TuMV) [24]. Macovei et al. [34] developed rice plants resistant to Rice tungro spherical virus (RTSV) through mutation of elF4G gene. Similarly, the disruption of the cucumber (Cucumis sativus) elF4E gene provided plant resistance to multiple members of the Potyviridae, namely the ipomovirus Cucumber vein yellowing virus (CVYV) and the potyviruses Zucchini yellow mosaic virus (ZYMV) and Papaya ringspot mosaic virus (PRSV) [32]. Resistance against Clover yellow vein virus (CYVV) was achieved in A. thaliana plants by targeting the elF4E1 gene using CRISPR/Cas9 [35]. Very recently, CRISPR/Cas9 has also allowed to perform double mutations on the novel cap-binding protein-1 and protein-2 (nCBP-1 and nCBP-2) belonging to the elF4E family, on cassava, which increased the resistance to Cassava brown streak virus (CBSV) [36].
Table 1. CRISPR/Cas for viral resistance in plants by targeting viral genome (virus mediated resistance) and host factors (plant mediated resistance).
| Plant Species | Target Virus | Type of Resistance | Targeting Genomic Regions |
|---|---|---|---|
| N. benthamiana | BeYDV | DNA virus mediated | sgRNAs targeting LIR and rep/RepA |
| A. thaliana and N. benthamiana | BSCTV | DNA virus mediated | 43 sgRNAs targeting BSCTV genome |
| N. benthamiana | TYLCV | DNA virus mediated | sgRNAs targeting Rep and CP |
| N. benthamiana | CLCuKoV | DNA virus mediated | sgRNAs targeting non-coding IR, CP and Rep |
| TYLCV | |||
| TYLCSV | |||
| MeMV | |||
| BCTV-Worland | |||
| BCTV-Logan | |||
| A. thaliana | TuMV | Plant mediated | elF(iso)4E knock out |
| Cucumis sativus | CVYV | Plant mediated | elF4E knock out |
| ZYMV | |||
| PRSV | |||
| A. thaliana | CYVV | Plant mediated | elF4E1 |
| Oryza sativa | RTSV | Plant mediated | elF4G knock out |
| A. thaliana | CaMV | DNA virus mediated | sgRNAs targeting CP |
| A. thaliana and N. benthamiana | CMV | RNA virus mediated | 22 sgRNAs targeting CMV genome and 3 sgRNAs targeting TMV genome |
| TMV | |||
| N. benthamiana | TuMV | RNA virus mediated | sgRNAs targeting HC-Pro and CP |
| Cassava | CBSV | Plant mediated | nCBP-1 and nCBP-2 (elF4E family) |
| Barley | WDV | DNA virus mediated | sgRNAs targeting MP, CP, Rep(Rep A and LIR |
| N. benthamiana and Oryza sativa | TMV | RNA virus mediated | sgRNAs targeting 5 regions in TMV, 3 in SRBDSV and 3 in RSMV |
| SRBDSV | |||
| RSMV | |||
| Banana (Gonja manjaya) | eBSV | DNA virus mediated | sgRNAs targeting ORF1, ORF2 and ORF3 |
| Potato (Solanum tuberosum) | PVY | RNA virus mediated | sgRNAs targeting P3, CI, NIb and CP |
It is a fact that modifications of plant genes may always face the risk to interfere with plant functions associated to those genes, with a fitness cost for the host, however these examples have demonstrated the success of CRISPR/Cas9 to produce genetic resistant plants through plant mediated resistance and without compromising plant functions.
2.2. CRISPR for Virus Mediated Resistance
Another approach to achieve plant viral resistance through CRISPR systems is by directly targeting viral genomes. In this approach, the problems that may arise by interfering with genes, that may also be associated to other plant functions—such as growth, reproduction, or others—are surpassed. However, for this type of mediated resistance, CRISPR/Cas systems must specifically directly target and cleave DNA of DNA viruses, or RNA of RNA viruses [25].
CRISPR for virus mediated resistance was first exploited to fight DNA viruses, as the discovery of CRISPR/Cas systems that can cleave RNA was more recent [8][20]. The discovery of such systems (class II, type VI Cas effectors, and Cas9 variants)—namely Cas13a (C2c2), Cas13b (C2c6), Cas13c (C2c7), Cas13d, FnCas9, and RCas9 (RNA targeting SpCas9) [1][8][37][38][39][40], was a great benefit—enabling direct targeting of RNA viruses which represent most plant pathogenic viruses.
Several studies have demonstrated the potential of CRISPR to impart plant resistance by targeting either DNA or RNA viral genomes, causing delayed or reduced accumulation of viruses and significantly attenuating symptoms of infection [41]. Some of those studies which directly mutate DNA and RNA viruses in plants expressing CRISPR/Cas machinery are described below (Table 1).
There are two major groups of plant DNA viruses, the double stranded caulimoviruses and the geminiviruses, the later which, although single stranded, replicate within the plant cell as double stranded DNA [42]. According to the latest report of the international Committee on Taxonomy of Viruses (ICTV), the Geminiviridae is the largest group of plant viruses, with 485 species [43]. Geminiviruses infect many economically important crops such as cassava, watermelon, squash, petunia, tobacco, pepper, potato, tomato, bean, soybean, cowpea, cotton, and others, leading to reduced crop yields worldwide [44][45]. Due to this reason, it is not surprising that most DNA virus mediated resistance studies have been applied to geminiviruses (Table 1). Ali et al. [46] used sgRNA molecules targeting coding (rep genes and coat proteins) and non-coding sequences (conserved intergenic region) of the Tomato yellow leaf curl virus (TYLCV) genome, that were delivered via Tobacco rattle virus (TRV) system into Nicotiana benthamiana plants expressing Cas9, causing a reduction of accumulation of viral DNA and reduction of symptoms in plants. A subsequent study using CRISPR/Cas9 system with a sgRNA targeting a conserved region in multiple begomoviruses (CLCuKoV, TYLCV, TYLCSV, MeMV, BCTV-Worland and BCTV-Logan), simultaneously mediated interference and showed that the targeting of viral non-coding, intergenic sequences was more efficient, limiting the generation of recovered viral variants that evade CRISPR-mediated immunity by reverting the induced mutations through NHEJ [22]. Other studies have achieved plant viral resistance through the expression of sgRNAs complementary to sequences either within Bean yellow dwarf virus (BeYDV), Wheat dwarf virus (WDV) or Beet severe curly top virus (BSCTV) genomes, which reduced virus accumulation and symptoms in plants overexpressing Cas9 such as N. benthamiana, barley, and A. thaliana [23][47][48]. Similarly, CRISPR/Cas9 allowed to obtain resistance against banana streak disease by targeting endogenous Banana streak virus (eBSV) sequences [49].
Plant resistance to a caulimovirus was achieved when Liu et al. [19] expressed multiple sgRNAs targeting the caulimovirus Cauliflower mosaic virus (CaMV) coat protein gene in Arabidopsis plants and 20 days after mechanical inoculation of the virus, 85–90% of the plants remained symptomless and showed no presence of CaMV.
Immunity against the RNA viruses Cucumber mosaic virus (CMV) and Tobacco mosaic virus (TMV) was achieved in N. benthamiana and A. thaliana transgenic plants expressing FnCas9 and a sgRNA complementary to viral genome delivered through a pCambia based vector [50]. Another study showed that N. benthamiana expressing Cas13a either transiently (using binary vector pK2WG7) or constitutively, and expressing crRNAs complementary to different Tulip mosaic virus (TuMV) genomic regions, delivered through TRV system, interfered with viral replication and spread [20]. CRISPR/Cas13a (LshCas13a) system showed to target and degrade genomic RNA of TMV in N. benthamiana plants and to confer resistance to Southern rice black-streaked dwarf virus (SRBSDV) and Rice stripe mosaic virus (RSMV) in rice plants [21]. Zhan et al. [51] showed that transgenic potato lines expressing Cas13a/sgRNA constructs targeting conserved coding regions of different Potato virus Y (PVY) strains allowed to confer broad spectrum resistance against multiple PVY strains.
As stated above, many studies have shown the great versatility of the CRISPR technology towards plant virus resistance and have successfully shown the production of viral resistant plants. CRISPR has the potential to accelerate viral resistance breeding, since it is more effective and rapid than conventional breeding. In addition, CRISPR has the capacity to target virus directly and therefore to be applied to crops with limited genome sequence information.
There are also limitations of the use of CRISPR in virus plant resistance that must not be discarded. Knocking out essential host factors may always lead to the possibility of plant lethality or impaired growth [52][53]. Although many studies concerning mutations of host factors did not report any negative effects, the introduction of point mutations in host factor genes, instead of knocking out, should be considered, so that it does not interfere with plant growth but still prevents viral infection [54]. Another important limitation of CRISPR is the undesirable genomic modifications of plant genome, the off-targets. Although much less common to occur in plants than in other systems, off-target mutations may be avoided by the use of catalytically inactive Cas nucleases [55] or by using systems that only target RNA, which will be further destroyed by the plant silencing system.
CRISPR/Cas requires the optimal selection of sgRNA target sites to ensure that targeted viruses do not evolve mutations that escape from CRISPR/Cas cleavage, and that novel and more severe strains that cannot be cleaved again do not arise [22][56]. Additionally, multiplex targeting and targeting noncoding regions of viral genomes have shown to reduce viral mutation rates and minimize the formation of new viral strains capable of infection [22]. Also, CRISPR/Cas systems that target or bind RNA can be used together with Cas9 to reduce the RNA intermediates of DNA viruses, eliminating the viruses that may escape the CRISPR/Cas9 machinery [22]. FnCas9 has shown binding capacity to viral transcripts which probably provides even more durable resistance than nucleases that provide direct targeting [25].
There is still a long way to go concerning the full potential of CRISPR/Cas systems for engineering plant virus resistance, and more studies still need to be performed to improve their efficiency. However, it is clear that CRISPR is a milestone in plant virus resistance and the utilization of this technology in agriculture will certainly result in higher yields and quality of plants.
This entry is adapted from the peer-reviewed paper 10.3390/v13010141
References
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017, 15, 169–182.
- Rath, D.; Amlinger, L.; Rath, A.; Lundgren, M. The CRISPR-Cas immune system: Biology, mechanisms and applications. Biochimie 2015, 117, 119–128.
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821.
- Wang, M.; Lu, Y.; Botella, J.R.; Mao, Y.; Hua, K.; Zhu, J. Gene Targeting by Homology-Directed Repair in Rice Using a Geminivirus-Based CRISPR/Cas9 System. Mol. Plant 2017, 10, 1007–1010.
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826.
- Yang, H.; Ren, S.; Yu, S.; Pan, H.; Li, T.; Ge, S.; Zhang, J.; Xia, N. Methods favoring homology-directed repair choice in response to crispr/cas9 induced-double strand breaks. Int. J. Mol. Sci. 2020, 21, 6461.
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2018, 551, 464–471.
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573.
- Gao, C. The future of CRISPR technologies in agriculture. Nat. Rev. Mol. Cell Biol. 2018, 19, 275–276.
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442.
- Khan, M.Z.; Amin, I.; Hameed, A.; Mansoor, S. CRISPR–Cas13a: Prospects for Plant Virus Resistance. Trends Biotechnol. 2018, 36, 1207–1210.
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 2014, 9, 2395–2410.
- Brooks, C.; Nekrasov, V.; Lipppman, Z.B.; Van Eck, J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014, 166, 1292–1297.
- Jia, H.; Nian, W. Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 2014, 9.
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 2017, 15, 1509–1519.
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.L.; Gao, C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617.
- Char, S.N.; Neelakandan, A.K.; Nahampun, H.; Frame, B.; Main, M.; Spalding, M.H.; Becraft, P.W.; Meyers, B.C.; Walbot, V.; Wang, K.; et al. An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol. J. 2017, 15, 257–268.
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017, 15, 207–216.
- Liu, H.; Soyars, C.L.; Li, J.; Fei, Q.; He, G.; Peterson, B.A.; Meyers, B.C.; Nimchuk, Z.L.; Wang, X. CRISPR/Cas9-mediated resistance to cauliflower mosaic virus. Plant Direct 2018, 2, e00047.
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018, 19, 1.
- Zhang, T.; Zhao, Y.; Ye, J.; Cao, X.; Xu, C.; Chen, B.; An, H.; Jiao, Y.; Zhang, F.; Yang, X.; et al. Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol. J. 2019, 17, 1185–1187.
- Ali, Z.; Ali, S.; Tashkandi, M.; Zaidi, S.S.E.A.; Mahfouz, M.M. CRISPR/Cas9-Mediated Immunity to Geminiviruses: Differential Interference and Evasion. Sci. Rep. 2016, 6, 26912.
- Ji, X.; Zhang, H.; Zhang, Y.; Wang, Y.; Gao, C. Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 2015, 1, 15144.
- Pyott, D.E.; Sheehan, E.; Molnar, A. Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 2016, 17, 1276–1288.
- Zaidi, S.S.E.A.; Tashkandi, M.; Mansoor, S.; Mahfouz, M.M. Engineering plant immunity: Using CRISPR/Cas9 to generate virus resistance. Front. Plant Sci. 2016, 7, 1673.
- Hull, R. Matthews’ Plant Virology, 4th ed.; Academic Press: New York, NY, USA, 2001.
- Hashimoto, M.; Neriya, Y.; Yamaji, Y.; Namba, S. Recessive resistance to plant viruses: Potential resistance genes beyond translation initiation factors. Front. Microbiol. 2016, 7, 1695.
- de Ronde, D.; Butterbach, P.; Kormelink, R. Dominant resistance against plant viruses. Front. Plant Sci. 2014, 5, 1–17.
- Wittmann, S.; Chatel, H.; Fortin, M.G.; Laliberté, J.F. Interaction of the viral protein genome linked of turnip mosaic potyvirus with the translational eukaryotic initiation factor (iso) 4E of Arabidopsis thaliana using the yeast two-hybrid system. Virology 1997, 234, 84–92.
- Cillo, F.; Palukaitis, P. Transgenic Resistance; Academic Press: New York, NY, USA, 2014; Volume 90, ISBN 9780128012468.
- Sanfaçon, H. Plant translation factors and virus resistance. Viruses 2015, 7, 3392–3419.
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Sherman, A.; Arazi, T.; Gal-On, A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 2016, 17, 1140–1153.
- Borrelli, V.M.G.; Brambilla, V.; Rogowsky, P.; Marocco, A.; Lanubile, A. The enhancement of plant disease resistance using crispr/cas9 technology. Front. Plant Sci. 2018, 9.
- Macovei, A.; Sevilla, N.R.; Cantos, C.; Jonson, G.B.; Slamet-Loedin, I.; Čermák, T.; Voytas, D.F.; Choi, I.R.; Chadha-Mohanty, P. Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 2018, 16, 1918–1927.
- Bastet, A.; Lederer, B.; Giovinazzo, N.; Arnoux, X.; German-Retana, S.; Reinbold, C.; Brault, V.; Garcia, D.; Djennane, S.; Gersch, S.; et al. Trans-species synthetic gene design allows resistance pyramiding and broad-spectrum engineering of virus resistance in plants. Plant Biotechnol. J. 2018, 16, 1569–1581.
- Gomez, M.A.; Lin, Z.D.; Moll, T.; Chauhan, R.D.; Hayden, L.; Renninger, K.; Beyene, G.; Taylor, N.J.; Carrington, J.C.; Staskawicz, B.J.; et al. Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 2019, 17, 421–434.
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027.
- Yan, W.X.; Chong, S.; Zhang, H.; Makarova, K.S.; Koonin, E.V.; Cheng, D.R.; Scott, D.A. Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Mol. Cell 2018, 70, 327–339.e5.
- O’Connell, M.R.; Oakes, B.L.; Sternberg, S.H.; East-Seletsky, A.; Kaplan, M.; Doudna, J.A. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 2014, 516, 263–266.
- Price, A.A.; Sampson, T.R.; Ratner, H.K.; Grakoui, A.; Weiss, D.S. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc. Natl. Acad. Sci. USA 2015, 112, 6164–6169.
- Mahas, A.; Mahfouz, M. Engineering virus resistance via CRISPR–Cas systems. Curr. Opin. Virol. 2018, 32, 1–8.
- van Regenmortel, M.H.V.; Fauquet, C.M.; Bishop, D.H.L.; Carstens, E.B.; Estes, M.K.; Lemon, S.M.; Maniloff, J.; Mayo, M.A.; McGeoch, D.J.; Pringle, C.R.; et al. Virus Taxonomy Seventh Report of the International Committee on Taxonomy of Viruses; Academic Press: San Diego, CA, USA, 2000; ISBN 0-12-370200-3.
- ICTV. Available online: (accessed on 14 September 2020).
- Moffat, A.S. Geminiviruses Emerge as Serious Crop Threat. Science 1999, 286, 1835.
- Zaidi, S.S.A.; Tashkandi, M.; Mahfouz, M.M.; Zaidi, S.S.A. Engineering Molecular Immunity Against Plant Viruses. Prog. Mol. Biol. Transl. Sci. 2017, 149, 167–186.
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015, 16.
- Baltes, N.J.; Hummel, A.W.; Konecna, E.; Cegan, R.; Bruns, A.N.; Bisaro, D.M.; Voytas, D.F. Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nat. Plants 2015, 1.
- Kis, A.; Hamar, É.; Tholt, G.; Bán, R.; Havelda, Z. Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnol. J. 2019, 17, 1004–1006.
- Tripathi, J.N.; Ntui, V.O.; Ron, M.; Muiruri, S.K.; Britt, A.; Tripathi, L. CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2019, 2, 46.
- Zhang, T.; Zheng, Q.; Yi, X.; An, H.; Zhao, Y.; Ma, S.; Zhou, G. Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol. J. 2018, 16, 1415–1423.
- Zhan, X.; Zhang, F.; Zhong, Z.; Chen, R.; Wang, Y.; Chang, L.; Bock, R.; Nie, B.; Zhang, J. Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol. J. 2019, 17, 1814–1822.
- Callot, C.; Gallois, J.L. Pyramiding resistances based on translation initiation factors in Arabidopsis is impaired by male gametophyte lethality. Plant Signal. Behav. 2014, 9.
- Gauffier, C.; Lebaron, C.; Moretti, A.; Constant, C.; Moquet, F.; Bonnet, G.; Caranta, C.; Gallois, J.L. A TILLING approach to generate broad-spectrum resistance to potyviruses in tomato is hampered by eIF4E gene redundancy. Plant J. 2016, 85, 717–729.
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788.
- Naeem, M.; Majeed, S.; Hoque, M.Z.; Ahmad, I. Latest Developed Strategies to Minimize the Off-Target Effects in CRISPR-Cas-Mediated Genome Editing. Cells 2020, 9, 1608.
- Mehta, D.; Stürchler, A.; Anjanappa, R.B.; Zaidi, S.S.E.A.; Hirsch-Hoffmann, M.; Gruissem, W.; Vanderschuren, H. Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol. 2019, 20, 80.
