Spondylocostal dysostosis (SCDO) is a rare heritable congenital condition, characterized by multiple severe malformations of the vertebrae and ribs.
- spondylocostal dysostosis
- somitogenesis clock
- somite formation
1. Introduction
Biomedical research employs multiple scientific inquiry approaches, such as basic, clinical, and translational research and great advances have been made in any one of these fronts. The ultimate aim is to have such a clear understanding of the molecular and cellular causes of the phenotypical and clinical manifestations of Human diseases, so that a successful therapeutic solution or preventive strategy may be developed. Developmental Biology has greatly contributed to understanding the pathophysiology of multiple congenital diseases. Congenital malformations, defined as structural or functional abnormalities that are present since birth, are globally responsible for 11% of newborn deaths [1]. These are largely due to genetic causes; however, phenotypic variability also depends on epigenetic components and environmental factors to which the individual is exposed during embryo development [2]. Within congenital malformations, spinal dysplasias occur in about 1 in 5000 births [3] and spondylocostal dysostosis (SCDO), considered a rare genetic disease, has an estimated incidence of 1 in every 40,000 births [4].
2. Spondylocostal Dysostosis
The term Spondylocostal dysostosis (SCDO) includes an etiologically heterogeneous group of heritable diseases characterized by congenital malformations of the axial skeleton at the level of the vertebrae and ribs, with varying degrees of severity. In 1938, Saul Jarcho and Paul M. Levin made the first description of what is currently classified as Type 2 SCDO [5]. For many years thereafter, the Jarcho-Levin Syndrome classification covered any individual with congenital defects in vertebrae segmentation and rib abnormalities, even if only remotely related to the syndrome originally described [6].
Phenotypically, SCDO patients are characterized by a compressed trunk that is disproportionately short relative to the individual’s total height, a short and rigid neck, and protrusion of the abdomen, often accompanied by moderate and non-progressive scoliosis [6]. Typically, segmentation defects occur in 10 contiguous vertebrae. Respiratory complications are among the most common causes of morbidity and mortality in affected individuals; the restrictions imposed by the deformation of the rib cage affect lung development and signs and symptoms such as tachypnea, fatigue or lung infections are often present as a result of pulmonary hypoplasia [7] and abnormal chest compliance [8]. SCDO usually occurs in isolation, with manifestation of abnormalities restricted to the spine and ribs [6]. A somewhat similar, albeit a clinically and radiologically independent disease, is spondylothoracic dysostosis (STD), first described by Lavy, Palmer, and Merritt in 1966 [4][9]. STD’s main distinguishing feature is a shorter but symmetrical chest, with fused thoracic vertebrae, and overall-normal ribs that are fused next to their vertebral origin acquiring a fan-like or crab-like configuration. STD is often associated with early death from respiratory dysfunction or infection [4][8].
The diagnosis of SCDO is based on the clinic and the radiological images. Radiological findings are diversified and include the involvement of several segments of the spine, especially in the thoracic region, which present butterfly-like vertebrae, fused hemivertebrae or vertebrae, and also diverse alterations to rib morphology (Figure 1). These include rib enlargement, fusion, bifurcation or even agenesis, causing asymmetries with incorrect rib alignment and often with a reduction in the total number of ribs [7]. The identification of the specific disease type is confirmed by genetic tests to identify mutant alleles in one of the seven currently known genes responsible for the different types of SCDO [6].
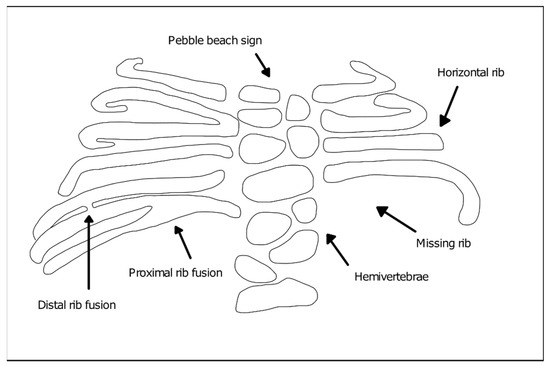
Figure 1. Vertebral and rib malformations identified in different types of spondylocostal dysostosis (SCDO).
Genetics and Main Phenotypic Characteristics of SCDO Subtypes
SCDO is a genetic disease with an autosomal inheritance pattern that is predominantly recessive [10]. The disease is most common in cases of consanguinity of the parents and is associated with the transmission of pathological mutations in genes with a key role in the formation of the axial skeleton during embryogenesis. The SCDO-associated genes known to date and their associated SCDO subtypes are the following:
-
DLL3 (Delta-like protein 3)—Type 1 SCDO (OMIM #277300). Type 1 SCDO is the most common form found in clinical practice and the majority of the affected individuals result from inbreeding unions [6]. DLL3 mutations identified in patients include insertions, frameshift, splicing, and nonsense mutations leading to premature truncation or protein function impairment [11][12]. Phenotypically, the mutation of this gene leads to moderate, non-progressive scoliosis and rarely requires surgical intervention to stabilize the spine. The affected individuals consistently show an irregular ossification pattern, with the vertebral bodies assuming a rounded or oval shape during childhood (“pebble beach sign”), evolving into irregular vertebral bodies and hemivertebrae as ossification is completed [6];
-
MESP2 (Mesoderm posterior protein 2)—Type 2 SCDO (OMIM #608681). The pathogenic variant of this gene results in straight ribs with fewer fusion points and therefore, more regularly aligned when compared to other types. SCDO type 2 has been described in three families, one of them with consanguineous parents [6]. Reported MESP2 missense mutations introduce premature stop codons leading to protein truncation [13]. Other mutations thought to severely reduce protein levels due to nonsense-mediated mRNA decay, are found in cases of STD and are associated with more severe phenotypes [6][13];
-
LFNG (Lunatic Fringe)—Type 3 SCDO (OMIM #609813). There are two reports of LFNG mutations associated with SCDO. The first documented individual presented a more severe shortening of the spine than that found in the other SCDO subtypes, with all vertebral bodies exhibiting severe segmentation defects. Rib anomalies were similar to those in SCDO type 1 and 2 [14][15]. An additional report was made of an individual carrying two distinct mutations in LFNG, with multiple vertebral defects along the entire spine [16][17]. In both reported cases, the identified missense mutations were found to impair Lfng enzymatic activity and/or subcellular localization [17]. Comparable vertebral malformations were described in individuals with mutations in the SLC35A3 gene encoding the Golgi UDP-GlcNAc transporter [18] (required for Lfng substrate availability), which further supports the importance of Lfng activity in axial skeleton formation;
-
HES7 (Hairy enhancer of split 7)—Type 4 SCDO (OMIM #613686). HES7 belongs to the family of hairy-enhancer-of-split transcription factors and is specifically expressed in the embryonic paraxial presomitic mesoderm (PSM) [19]. Mutations in HES7 were described in infants presenting a shortened spine, with segmentation defects predominantly in the thoracic region and irregularly aligned, fused ribs [20][21]. The identified missense mutations resulted in significant reduction of HES7 transcriptional inhibitory activity and alterations to its heterodimerization potential [20][21]. In some cases, neural tube closure defects were also present, although there is no evidence of a direct association of the two conditions. Type 4 SCDO was described in three families, one of which also had inbreeding [6];
-
TBX6 (T-box transcription factor 6)—Type 5 SCDO (OMIM #122600). SCDO-associated mutations in TBX6 were described in three generations of the same family, following an autosomal dominant inheritance. The affected individuals, all male, had a mixture of hemivertebrae and blocks of fused vertebral segments, moderate scoliosis affecting the middle thoracic region, with little involvement of the ribs [22][23]. Multiple other cases have also been reported and the underlying TBX6 mutations include 16p11.2 genomic deletions, as well as nonsense and frameshift mutations, some of which were found to alter TBX6 subcellular localization and/or transcriptional activity [16][24][25];
-
RIPPLY2 (Protein ripply 2)—Type 6 SCDO (OMIM #616566). The first report of mutations in this gene described two brothers who had vertebral segmentation defects in cervical and thoracic regions, including hemivertebrae and butterfly vertebrae but overall normal ribs, with marked cervical kyphosis and moderate thoracic scoliosis [6][26]. One of the reported mutations introduces a premature stop codon, with consequent loss of transcriptional repressor activity; the other is a missense mutation localized at a mRNA splice site, but its functional consequences have not yet been elucidated [26]. Since then, RIPPLY2 mutations were described in several other individuals with vertebral defects, many times associated with additional congenital malformations [27][28];
-
DMRT2 (Doublesex And Mab-3 Related Transcription Factor 2; OMIM *604935). An homozygous DMRT2 variant, predicted to lead to the absence of full length DMRT2 protein product due to loss of the start codon, was recently associated with a severe form of a SCDO-like phenotype [29]. The newborn presented severe costovertebral defects, with all ribs affected either in size or shape (missing, fused, bifid, and hypoplastic), particularly in the most distal part. The vertebrae were also malformed (laminae intervertebral fusions and irregular ossification), despite the absence of clear segmentation defects of the vertebral bodies.
Despite the growing knowledge of the clinical and genetic aspects associated with SCDO, significant challenges still remain in understanding the underlying pathological mechanisms. Developmental engineering is a promising area in terms of treatment strategies for diseases related to embryonic development of the spine. Its success however depends on the ever-growing knowledge of the disease’s embryonic etiology together with the embryonic developmental processes affected [30].
3. Formation of the Spine during Embryogenesis
The human axial skeleton is an extraordinarily structured system that starts to form early in embryo development. Soon after blastocyst implantation in the uterus, gastrulation takes place giving rise to the three embryonic germ layers: ectoderm, mesoderm, and endoderm. The PSM, located bilaterally to the axial neural tube, is then segmented into round-shaped transient structures called somites. As new somite pairs are periodically formed in the anterior region, new cells are added to the posterior PSM, allowing somitogenesis to progress simultaneously with axial extension (reviewed in [31][32]). Between the 20th day and the 5th week of gestation, a total of 42 to 44 pairs of somites are formed, with each pair of somites appearing every 5 h [33][34]. Temporal precision of somitogenesis is observed in all vertebrates, with species-specific periodicities, such as 90 min in the chicken and 2 h in the mouse embryo (reviewed in [31]). Later, somites will differentiate into vertebrae, ribs, and the skeletal muscles that provide stability and movement to the axial skeleton.
3.1. The Somitogenesis Molecular Clock
The temporal and spatial precision of somitogenesis has long intrigued many scientists. One of the first theoretical models to explain how this process is regulated—the Clock and Wavefront model—proposed the existence of a molecular oscillator (Clock) that would define the tempo of somite formation and of a Wavefront of cell differentiation, that would set the position of each new somite boundary [35]. The experimental evidence of such a molecular oscillator was provided 20 years later, when Palmeirim et al. described hairy1 gene expression oscillations in the chicken PSM with the same periodicity as somite formation, 90 min [36] (Figure 2). Since then, many genes with oscillatory expression in the PSM were found in multiple vertebrate species, suggesting that this is a conserved feature of vertebrate embryos [37]. Strikingly, the clock periodicity in each species is directly coupled to the time required to form a pair of somites. This is 90 min for the chicken embryo, 2 h in the mouse, and approximately 5 h in humans.
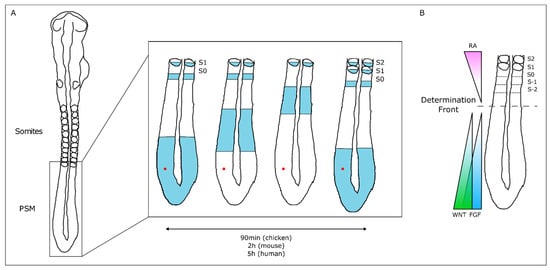
Figure 2. The Molecular Clock and Signaling Gradients in temporal/spatial control of vertebrate somitogenesis. (A) Representation of one oscillation of hairy1 gene expression (blue) in the presomitic mesoderm (PSM) of a 48h chicken embryo. A cell in the PSM (red dot) undergoes a complete cycle of gene expression activation-repression-activation as a new somite is formed in the anterior-most PSM. (B) Opposing gradients of retinoic acid (RA) and WNT/FGF signaling converge at the determination front; rostrally, the PSM tissue is already committed to form somites.
The wavefront proposed in the Clock and Wavefront model [35] plays an important role in the correct positioning of each new somite boundary in the anterior-end of the PSM [38]. An anterior retinoic acid (RA) signaling gradient opposes posterior-derived Fgf and Wnt gradients, establishing the so-called determination front, where PSM cells become irreversibly committed to form a somite within the next 3–4 molecular clock oscillations [38][39][40]. Posterior-to-anterior gradients of Wnt and Fgf signaling activity maintain cells in an undifferentiated state in the posterior PSM. Overexpressing Fgf8 at the level of the determination front leads to the formation of smaller somites, while if Fgf signaling is inhibited, larger somites are formed [38]. Moreover, both Fgf and Wnt signaling are crucial for embryo elongation, thus coupling the process of somitogenesis with anterior-to-posterior elongation [41][42].
Somitogenesis clock gene expression oscillations are cell autonomous, maintained by negative feedback loops (Figure 3) (reviewed in [23]). The sum of the temporal delays involved in the process—from transcription, RNA splicing and decay, to protein translation and degradation—is crucial for robust oscillations to occur with a defined rhythm [31]. The deletion of all introns in mouse Hes7, for example, abolished oscillatory expression dynamics, causing sustained Hes7 expression in the PSM and severe vertebral defects [43]. Also, a reduction in the number of Hes7 introns led to accelerated oscillations and supernumerary somites, further evidencing the importance of the temporal delays imposed by mRNA splicing in periodic clock oscillations [44]. Another important feature is the mRNA and protein degradation rates. Nitanda and collaborators (2014) showed that the 3′UTR of clock mRNAs is a key defining feature for their expression patterns. By exchanging the 3′UTRs of Hes7 and Lfng, they found that the expression patterns obtained were dictated by the 3′UTR sequence per se, and not the gene it originally belonged to [45]. 3′UTRs are important to regulate the half-life of the respective mRNAs, partly because these regions are targeted by microRNAs, which can affect both the period and amplitude of the oscillations, or even lead to dampening and loss of the oscillatory dynamics [46][47]. The clock proteins’ half-life also plays an important role in the maintenance of the oscillatory dynamics [48]. Hirata and collaborators (2004) showed that increasing Hes7 half-life from ~20 to ~30 min lead to progressive dampening and loss of oscillations and, consequently, to somite segmentation defects [48]. Interestingly, it was recently shown that the differences in cell-autonomous oscillation periods of the segmentation clock between species can be explained by the difference in the kinetics of the biochemical reactions involved in the different time delays, and not by the gene sequences themselves [49].
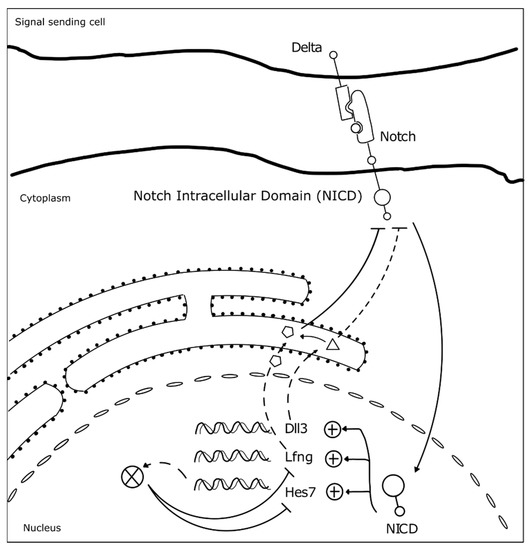
Figure 3. Negative feedback loops underlie somitogenesis clock oscillations. The identified mutated genes in SCDO are represented. Upon Notch receptor activation by a Delta ligand from a neighbor cell, the Notch intracellular domain (NICD) translocates to the nucleus where it activates the transcription of HES7, LFNG and DLL3 [23]. Lfng and Dll3 act cooperatively to repress further NICD activation [50], while Hes7 acts to represses its own expression and that of Lfng [23].
At the tissue level, the molecular oscillator needs to be synchronized between neighboring cells, in order to allow the correct number of cells to simultaneously differentiate into a new somite. Work performed in mouse and zebrafish embryos showed that Delta-Notch signaling is required to couple oscillations between adjacent cells [51][52][53]. Additionally, oscillations of genes belonging to different signaling pathways need to be coupled. In fact, many clock genes are components of the Fgf, Wnt, and Notch signaling pathways [54][55], evidencing that the somitogenesis molecular clock is a complex network of oscillators, connected to ensure proper morphogenesis. For example, activation of Hes7 expression in the embryo tail bud is dependent on Fgf and not Notch signaling, which is then required to maintain Hes7 expression throughout the PSM. Hes7 in turn activates Fgf signaling modulators, such as the negative regulator Dusp4 [51]. Wnt signaling-dependent Axin2 also oscillates in the posterior PSM, out-of-phase relative to Lfng and Hes7 [39]. The coupling between Notch, Fgf, and Wnt signaling pathways in somitogenesis was beautifully evidenced by performing experiments with microfluidics devices, which showed that when Notch is entrained, Wnt expression dynamics is altered accordingly [56]. This is particularly important in the anterior PSM where the signaling pathways must converge to the same phase for proper somite segmentation [56].
Embryo clock synchronization between the left and right PSMs is crucial to ensure that somite segmentation occurs in a bilateral symmetric fashion. In zebrafish embryos Terra/Dmrt2a was shown to be necessary to maintain symmetrical clock gene expression patterns, as well as the same number of somites, in the left-right PSMs [57]. This role in left-right patterning is not conserved in the mouse embryo, but Dmrt2 was found to be indispensable for proper somite maturation [58] and formation of the vertebrae and ribs [59].
3.2. Somite Segmentation and Vertebrae Formation
PSM cells located posteriorly to the determination front are under the influence of high levels of Wnt and Fgf signaling and express the paraxial mesoderm marker Tbx6 (Figure 4). As the embryo elongates and cells in the anterior PSM leave the Fgf8 domain, Tbx6 and Notch are able to activate Mesp2 expression, delimiting in the caudal boundary of the next somite to be formed. Mesp2 plays a central role in the formation of the somitic boundary (reviewed in [60]). Mesp2 activates Ripply2 expression, which then represses Tbx6 [60][61]. Mesp2 also activates EphA4 which in turn induces the expression of EphrinB2 rostrally, in the caudal region of the somite-to-be. Here, EphrinB2 initiates a molecular cascade leading to rearrangements in the extracellular matrix [62] and to a mesenchymal-to-epithelial (MET) transition, culminating in somite boundary formation. EphA and EphrinB2 are then restricted to the rostral and caudal portion of the somite, respectively, ensuring proper somite polarity [63] (Figure 4).
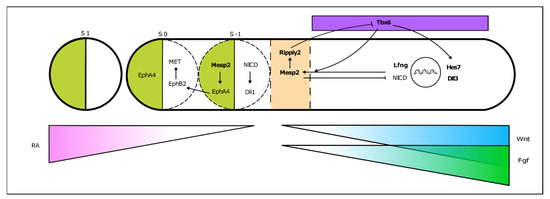
Figure 4. From somitogenesis clock oscillations to somite boundary formation. Periodic activation of Mesp2 at the rostral PSM, where FGF signaling is greatly decreased, triggers a molecular cascade ultimately leading to the budding off of a new somite. This is a simplified schematic representation, where the identified mutated genes in SCDO are highlighted.
As somitogenesis progresses, the newly formed epithelial somites will undergo modifications and differentiate into multiple structures. Initially, the medial-ventral portion of the somite undergoes an epithelial-to-mesenchymal transition under the influence of Shh signaling emanating from the notochord and neural tube floor plate. This is now the sclerotome, containing the precursors of the vertebrae and ribs (Figure 5). The dorsal-lateral portion of the somite remains epithelial and expresses Pax3, under the influence of Wnt signaling from the dorsal neural tube—the dermomyotome. The latter then further differentiates into the myotome, which will originate the axial musculature and the dermatome, which will form the dermis of the back [32]. Pax3 activates Dmrt2/Terra expression in the dermomyotome [64], where it plays an important role in somite patterning and myogenesis [59][64].
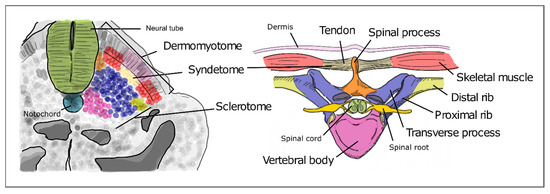
Figure 5. Differentiation of the different somite regions into definitive axial structures. After budding off from the PSM as epithelial spheres, somite differentiation takes place distinguishing the prospective cell populations that will give rise to adult structures. Colors in the figure represent precursor cells in the somite (left) and their respective definitive structures (right).
The sclerotome undergoes a re-segmentation process essential for the final configuration of the vertebrae and all the structures of the spine. After re-segmentation, the caudal segment of a sclerotome joins the rostral segment of the adjacent sclerotome to form a vertebra. Due to this process, each segment of the adult axial skeleton is derived from a caudal and rostral part of two consecutive somites instead of one somite. The dermomyotome maintains the initial segmented configuration of the somites, allowing the derived muscles to intercalate each skeletal segment. This is crucial for the mobility and stability of the axial skeleton in humans. The most ventral portion of the sclerotome then surrounds the notochord to give rise to the body of the vertebrae and the intervertebral discs. The notochord gives rise to the nucleus pulposus of the intervertebral discs. The vertebral arch originates from the sclerotome cells that surround the neural tube. The ribs originate from the sclerotome cells located in the ventro-lateral region (Figure 5) [65][66].
4. EVIDENCE FOR THE EMBRYONIC ORIGIN OF SCDOAs described above, SCDO patients present genetic mutations in genes involved in the somitogenesis molecular clock - DLL3, LFNG and HES7 - and in the formation of the somite boundaries - MESP2, TBX6 and RIPPLY2. Each mutated gene is at the origin of different types of SCDO (described above). An additional mutation in DMRT2 has been reported associated with a phenotype that could represent a new type of SCDO [29]. Clues to how mutations in these genes lead to SCDO and other congenital malformations arise from Developmental Biology studies, where the role of these genes has been extensively studied in the context of vertebrate embryo body segmentation. Mutant mice experimental models have greatly contributed to understanding the role of specific genes in this process and allow to study in further detail the molecular mechanisms that drive the onset of axial skeleton malformations. In fact, mice mutated in each one of the known genes that drive SCDO mostly recapitulate the phenotypes observed in Human SCDO patients, supporting a causal role for these mutations in the morphological malformations observed.
This entry is adapted from the peer-reviewed paper 10.3390/jdb9010005
References
- UNICEF; WHO; World Bank. UN DESA Levels & Trends in Child Mortality 2019. UN IGME Rep. 2019, 52.
- Krauss, R.S.; Hong, M. Gene–environment interactions and the etiology of birth defects. Curr. Top. Dev. Biol. 2016, 116, 569–580.
- Geister, K.A.; Camper, S.A. Advances in Skeletal Dysplasia Genetics. Annu. Rev. Genomics Hum. Genet. 2015, 16, 199–227.
- Berdon, W.E.; Lampl, B.S.; Cornier, A.S.; Ramirez, N.; Turnpenny, P.D.; Vitale, M.G.; Seimon, L.P.; Cowles, R.A. Clinical and radiological distinction between spondylothoracic dysostosis (Lavy-Moseley syndrome) and spondylocostal dysostosis (Jarcho-Levin syndrome). Pediatr. Radiol. 2011, 41, 384–388.
- Jarcho, S. Hereditary malformation of the vertebral bodies. Bull. Johns Hopkins Hosp. 1938, 62, 216–226.
- Turnpenny, P.D.; Sloman, M.; Dunwoodie, S.; ICVS (International Consortium for Vertebral Anomalies and Scoliosis). Spondylocostal Dysostosis, Autosomal Recessive. In GeneReviews [Internet]; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Stephens, K., Amemiya, A., Eds.; University of Washington Press: Seattle, WA, USA, 2017. Available online: (accessed on 24 January 2021).
- Southam, B.R.; Schumaier, A.P.; Crawford, A.H. Spondylocostal Dysostosis: A Literature Review and Case Report with Long-Term Follow-Up of a Conservatively Managed Patient. Case Rep. Orthop. 2018, 2018, 1–6.
- Campbell, R.M.J. Spine Deformities in Rare Congenital Syndromes: Clinical Issues. Spine 2009, 34, 1815–1827.
- Lavy, N.W.; Palmer, C.G.; Merritt, A.D. A syndrome of bizarre vertebral anomalies. J. Pediatrics 1966, 69, 1121–1125.
- Beine, O.; Bolland, J.; Verloes, A.; Lebrun, F.; Khamis, J.; Muller, C. Dysostosis spondylo-costale: It unites maladie génétique rare. Rev. Medicale Liege 2004, 59, 513–516.
- Bulman, M.P.; Kusumi, K.; Frayling, T.M.; McKeown, C.; Garrett, C.; Lander, E.S.; Krumlauf, R.; Hattersley, A.T.; Ellard, S.; Turnpenny, P.D. Mutations in the human delta homologue, DLL3, cause axial skeletal defects in spondylocostal dysostosis. Nat. Genet. 2000, 24, 438–441.
- Turnpenny, P.D.; Whittock, N.; Duncan, J.; Dunwoodie, S.; Kusumi, K.; Ellard, S. Novel mutations in DLL3, a somitogenesis gene encoding a ligand for the Notch signalling pathway, cause a consistent pattern of abnormal vertebral segmentation in spondylocostal dysostosis. J. Med. Genet. 2003, 40, 333–339.
- Whittock, N.V.; Sparrow, D.B.; Wouters, M.A.; Sillence, D.; Ellard, S.; Dunwoodie, S.L.; Turnpenny, P.D. Mutated MESP2 causes spondylocostal dysostosis in humans. Am. J. Hum. Genet. 2004, 74, 1249–1254.
- Sparrow, D.B.; Chapman, G.; Wouters, M.A.; Whittock, N.V.; Ellard, S.; Fatkin, D.; Turnpenny, P.D.; Kusumi, K.; Sillence, D.; Dunwoodie, S.L. Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am. J. Hum. Genet. 2006, 78, 28–37.
- Shifley, E.T.; VanHorn, K.M.; Perez-Balaguer, A.; Franklin, J.D.; Weinstein, M.; Cole, S.E. Oscillatory lunatic fringe activity is crucial for segmentation of the anterior but not posterior skeleton. Development 2008, 135, 899–908.
- Otomo, N.; Takeda, K.; Kawai, S.; Kou, I.; Guo, L.; Osawa, M.; Alev, C.; Kawakami, N.; Miyake, N.; Matsumoto, N.; et al. Bi-allelic loss of function variants of TBX6 causes a spectrum of malformation of spine and rib including congenital scoliosis and spondylocostal dysostosis. J. Med. Genet. 2019, 56, 622–628.
- Otomo, N.; Mizumoto, S.; Lu, H.F.; Takeda, K.; Campos-Xavier, B.; Mittaz-Crettol, L.; Guo, L.; Takikawa, K.; Nakamura, M.; Yamada, S.; et al. Identification of novel LFNG mutations in spondylocostal dysostosis. J. Hum. Genet. 2019, 64, 261–264.
- Edmondson, A.C.; Bedoukian, E.C.; Deardorff, M.A.; McDonald-McGinn, D.M.; Li, X.; He, M.; Zackai, E.H. A human case of SLC35A3-related skeletal dysplasia. Am. J. Med. Genet. A 2017, 173, 2758–2762.
- Bessho, Y.; Sakata, R.; Komatsu, S.; Shiota, K.; Yamada, S.; Kageyama, R. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 2001, 15, 2642–2647.
- Sparrow, D.B.; Guillén-Navarro, E.; Fatkin, D.; Dunwoodie, S.L. Mutation of HAIRY-AND-ENHANCER-OF-SPLIT-7 in humans causes spondylocostal dysostosis. Hum. Mol. Genet. 2008, 17, 3761–3766.
- Sparrow, D.B.; Sillence, D.; Wouters, M.A.; Turnpenny, P.D.; Dunwoodie, S.L. Two novel missense mutations in HAIRY-AND-ENHANCER-OF-SPLIT-7 in a family with spondylocostal dysostosis. Eur. J. Hum. Genet. 2010, 18, 674–679.
- Sparrow, D.B.; McInerney-Leo, A.; Gucev, Z.S.; Gardiner, B.; Marshall, M.; Leo, P.J.; Chapman, D.L.; Tasic, V.; Shishko, A.; Brown, M.A.; et al. Autosomal dominant spondylocostal dysostosis is caused by mutation in TBX6. Hum. Mol. Genet. 2013, 22, 1625–1631.
- Chen, W.; Liu, J.; Yuan, D.; Zuo, Y.; Liu, Z.; Liu, S.; Zhu, Q.; Qiu, G.; Huang, S.; Giampietro, P.F.; et al. Progress and perspective of TBX6 gene in congenital vertebral malformations. Oncotarget 2016, 7, 57430–57441.
- Wu, N.; Ming, X.; Xiao, J.; Wu, Z.; Chen, X.; Shinawi, M.; Shen, Y.; Yu, G.; Liu, J.; Xie, H.; et al. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N. Engl. J. Med. 2015, 372, 341–350.
- Takeda, K.; Kou, I.; Kawakami, N.; Iida, A.; Nakajima, M.; Ogura, Y.; Imagawa, E.; Miyake, N.; Matsumoto, N.; Yasuhiko, Y.; et al. Compound Heterozygosity for Null Mutations and a Common Hypomorphic Risk Haplotype in TBX6 Causes Congenital Scoliosis. Hum. Mutat. 2017, 38, 317–323.
- McInerney-Leo, A.M.; Sparrow, D.B.; Harris, J.E.; Gardiner, B.B.; Marshall, M.S.; O’Reilly, V.C.; Shi, H.; Brown, M.A.; Leo, P.J.; Zankl, A.; et al. Compound heterozygous mutations in RIPPLY2 associated with vertebral segmentation defects. Hum. Mol. Genet. 2015, 24, 1234–1242.
- Serey-Gaut, M.; Scala, M.; Reversade, B.; Ruaud, L.; Cabrol, C.; Musacchia, F.; Torella, A.; Accogli, A.; Escande-Beillard, N.; Langlais, J.; et al. Congenital posterior cervical spine malformation due to biallelic c.240-4T>G RIPPLY2 variant: A discrete entity. Am. J. Med. Genet. Part A 2020, 182, 1466–1472.
- Wegler, M.; Roth, C.; Schumann, E.; Kogan, J.; Totten, E.; Guillen Sacoto, M.J.; Abou Jamra, R.; Hornemann, F. Congenital cervical spine malformation due to bi-allelic RIPPLY2 variants in spondylocostal dysostosis type 6. Clin. Genet. 2021.
- Bouman, A.; Waisfisz, Q.; Admiraal, J.; van de Loo, M.; van Rijn, R.R.; Dimitra, M.; Oostra, R.J.; Mathijssen, I.B. Homozygous DMRT2 variant associates with severe rib malformations in a newborn. Am. J. Med. Genet. Part A 2018, 176, 1216–1221.
- Williams, S.; Alkhatib, B.; Serra, R. Development of the axial skeleton and intervertebral disc. Curr. Top. Dev. Biol. 2019, 133, 49–90.
- Pourquié, O. Segmentation of the vertebrate spine: From clock to scoliosis. Cell 2012, 145, 650–663.
- Gibb, S.; Maroto, M.; Dale, J.K. The segmentation clock mechanism moves up a notch. Trends Cell Biol. 2010, 20, 593–600.
- Kageyama, R.; Niwa, Y.; Isomura, A.; González, A.; Harima, Y. Oscillatory gene expression and somitogenesis. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 629–641.
- Chu, L.-F.; Mamott, D.; Ni, Z.; Bacher, R.; Liu, C.; Swanson, S.; Kendziorski, C.; Stewart, R.; Thomson, J.A. An In Vitro Human Segmentation Clock Model Derived from Embryonic Stem Cells. Cell Rep. 2019, 28, 2247–2255.
- Cooke, J.; Zeeman, E.C. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J. Theor. Biol. 1976, 58, 455–476.
- Palmeirim, I.; Henrique, D.; Ish-Horowicz, D.; Pourquié, O. Avian hairy Gene Expression Identifies a Molecular Clock Linked to Vertebrate Segmentation and Somitogenesis. Cell 1997, 91, 639–648.
- Pourquie, O. The vertebrate segmentation clock. J. Anat. 2001, 199, 169–175.
- Dubrulle, J.; McGrew, M.J.; Pourquié, O. FGF signaling controls somite boundary position and regulates segmentation clock control of spatiotemporal Hox gene activation. Cell 2001, 106, 219–232.
- Aulehla, A.; Wehrle, C.; Brand-Saberi, B.; Kemler, R.; Gossler, A.; Kanzler, B.; Herrmann, B.G. Wnt3a Plays a Major Role in the Segmentation Clock Controlling Somitogenesis. Dev. Cell 2003, 4, 395–406.
- Diez del Corral, R.; Storey, K.G. Opposing FGF and retinoid pathways: A signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. Bioessays 2004, 26, 857–869.
- Boulet, A.M.; Capecchi, M.R. Signaling by FGF4 and FGF8 is required for axial elongation of the mouse embryo. Dev. Biol. 2012, 371, 235–245.
- Nakaya, M.; Biris, K.; Tsukiyama, T.; Jaime, S.; Rawls, J.A.; Yamaguchi, T.P. Wnt3alinks left-right determination with segmentation and anteroposterior axis elongation. Development 2005, 132, 5425–5436.
- Takashima, Y.; Ohtsuka, T.; González, A.; Miyachi, H.; Kageyama, R. Intronic delay is essential for oscillatory expression in the segmentation clock. Proc. Natl. Acad. Sci. USA 2011, 108, 3300–3305.
- Harima, Y.; Takashima, Y.; Ueda, Y.; Ohtsuka, T.; Kageyama, R. Accelerating the tempo of the segmentation clock by reducing the number of introns in the Hes7 gene. Cell Rep. 2013, 3, 1–7.
- Nitanda, Y.; Matsui, T.; Matta, T.; Higami, A.; Kohno, K.; Nakahata, Y.; Bessho, Y. 3′–UTR-dependent regulation of mRNA turnover is critical for differential distribution patterns of cyclic gene mRNAs. FEBS J. 2014, 281, 146–156.
- Bonev, B.; Stanley, P.; Papalopulu, N. MicroRNA-9 Modulates Hes1 ultradian oscillations by forming a double-negative feedback loop. Cell Rep. 2012, 2, 10–18.
- Fujino, Y.; Yamada, K.; Sugaya, C.; Ooka, Y.; Ovara, H.; Ban, H.; Akama, K.; Otosaka, S.; Kinoshita, H.; Yamasu, K.; et al. Deadenylation by the CCR4-NOT complex contributes to the turnover of hairy-related mRNAs in the zebrafish segmentation clock. FEBS Lett. 2018, 592, 3388–3398.
- Hirata, H.; Bessho, Y.; Kokubu, H.; Masamizu, Y.; Yamada, S.; Lewis, J.; Kageyama, R. Instability of Hes7 protein is crucial for the somite segmentation clock. Nat. Genet. 2004, 36, 750–754.
- Matsuda, M.; Hayashi, H.; Garcia-Ojalvo, J.; Yoshioka-Kobayashi, K.; Kageyama, R.; Yamanaka, Y.; Ikeya, M.; Toguchida, J.; Alev, C.; Ebisuya, M. Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science 2020, 369, 1450–1455.
- Chapman, G.; Sparrow, D.B.; Kremmer, E.; Dunwoodie, S.L. Notch inhibition by the ligand DELTA-LIKE 3 defines the mechanism of abnormal vertebral segmentation in spondylocostal dysostosis. Hum. Mol. Genet. 2011, 20, 905–916.
- Niwa, Y.; Masamizu, Y.; Liu, T.; Nakayama, R.; Deng, C.X.; Kageyama, R. The Initiation and Propagation of Hes7 Oscillation Are Cooperatively Regulated by Fgf and Notch Signaling in the Somite Segmentation Clock. Dev. Cell 2007, 13, 298–304.
- Soza-Ried, C.; Öztürk, E.; Ish-Horowicz, D.; Lewis, J. Pulses of Notch activation synchronise oscillating somite cells and entrain the zebrafish segmentation clock. Development 2014, 141, 1780–1788.
- Yoshioka-Kobayashi, K.; Matsumiya, M.; Niino, Y.; Isomura, A.; Kori, H.; Miyawaki, A.; Kageyama, R. Coupling delay controls synchronized oscillation in the segmentation clock. Nature 2020, 580, 119–123.
- Dequéant, M.L.; Glynn, E.; Gaudenz, K.; Wahl, M.; Chen, J.; Mushegian, A.; Pourquié, O. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science 2006, 314, 1595–1598.
- Krol, A.J.; Roellig, D.; Dequéant, M.L.; Tassy, O.; Glynn, E.; Hattem, G.; Mushegian, A.; Oates, A.C.; Pourquié, O. Evolutionary plasticity of segmentation clock networks. Development 2011, 138, 2783–2792.
- Sonnen, K.F.; Lauschke, V.M.; Uraji, J.; Falk, H.J.; Petersen, Y.; Funk, M.C.; Beaupeux, M.; François, P.; Merten, C.A.; Aulehla, A. Modulation of Phase Shift between Wnt and Notch Signaling Oscillations Controls Mesoderm Segmentation. Cell 2018, 172, 1079–1090.e12.
- Saúde, L.; Lourenço, R.; Gonçalves, A.; Palmeirim, I. Terra Is a Left-Right Asymmetry Gene Required for Left-Right Synchronization of the Segmentation Clock. Nat. Cell Biol. 2005, 7, 918–920.
- Lourenço, R.; Lopes, S.S.; Saúde, L. Left-Right Function of dmrt2 Genes Is Not Conserved between Zebrafish and Mouse. PLoS ONE 2011, 5, 1–8.
- Seo, K.W.; Wang, Y.; Kokubo, H.; Kettlewell, J.R.; Zarkower, D.A.; Johnson, R.L. Targeted disruption of the DM domain containing transcription factor Dmrt2 reveals an essential role in somite patterning. Dev. Biol. 2006, 290, 200–210.
- Saga, Y. The mechanism of somite formation in mice. Curr. Opin. Genet. Dev. 2012, 22, 331–338.
- Zhao, W.; Ajima, R.; Ninomiya, Y.; Saga, Y. Segmental border is defined by Ripply2-mediated Tbx6 repression independent of Mesp2. Dev. Biol. 2015, 400, 105–117.
- Rifes, P.; Thorsteinsdóttir, S. Extracellular matrix assembly and 3D organization during paraxial mesoderm development in the chick embryo. Dev. Biol. 2012, 368, 370–381.
- Maroto, M.; Bone, R.A.; Kim Dale, J. Somitogenesis. Development 2012, 139, 2453–2456.
- Sato, T.; Rocancourt, D.; Marques, L.; Thorsteinsdóttir, S.; Buckingham, M. A Pax3/Dmrt2/Myf5 Regulatory Cascade Functions at the Onset of Myogenesis. PLoS Genet. 2010, 6, e1000897.
- Fleming, A.; Kishida, M.G.; Kimmel, C.B.; Keynes, R.J. Building the backbone: The development and evolution of vertebral patterning. Development 2015, 142, 1733–1744.
- Keynes, R. Patterning spinal nerves and vertebral bones. J. Anat. 2018, 232, 534–539.
