Sideroflexins (SLC56 family) are highly conserved multi-spanning transmembrane proteins inserted in the inner mitochondrial membrane in eukaryotes.
- sideroflexin
- mitochondria
- mitochondrial transporters
- iron homeostasis
- iron-sulfur cluster
1. Sideroflexins: From Structure to Function
1.1. Sideroflexins from an Historical Point of View
The mitochondrion is at the crossroad of key metabolic pathways (energy metabolism, central carbon metabolism, one carbon metabolism, lipid, nucleotides and amino acids synthesis, etc.) and is a key player in cell fate and response to stress or infection. In order to ensure its essential functions within the cell, the mitochondrion requires a wide variety of enzymes and transporters. Among these proteins, sideroflexins (SFXN) form a family of recently discovered mitochondrial proteins whose cell functions are progressively being specified. The first mention of the name “sideroflexin” appeared in 2001 [1]. Since then, a few studies have been dedicated to SFXN proteins, and at the time we were writing this review, only 24 articles were retrieved in Pubmed using the keyword “sideroflexin”. Pioneers in the SFXN field, Fleming et al. identified a mutation affecting the Sfxn1 gene in the flexed-tail mouse and proposed that the loss of Sfxn1 was responsible for the sideroblastic anemia phenotype. Thus, SFXN owe their name to the mice in which they were discovered (SIDEROblastic anemia and FLEXed-tail mouse) [1]. However, it should be noticed that the causal link between the mutation in the Sfxn1 gene and the phenotype of flexed-tail mice has not been clearly established yet. It was even questioned following a study showing that flexed tailed mice also had a mutation of the Madh5/Smad5 gene, involved in the BMP pathway, which could explain the anemia and flexed-tail phenotype [2][3].
1.2. The Sideroflexin Family: From Genes to Proteins
Sideroflexins (forming the SFXN/SLC56 family of mitochondrial transporters [4]) are highly conserved throughout eukaryotes. Only one sideroflexin is found in yeast (Fsf1 for Fungal sideroflexin 1), whereas there are two SFXNs in Drosophila (dSfxn1/3 and dSfxn2) and five SFXN (SFXN1-5) in vertebrates [1][5][6][7]. Our purpose is not to give an extensive overview of SFXN tissue distribution in this review, but some data are available in the literature. For example, SFXN1 mRNA levels in normal tissues and human cancers, as well as tissue distribution of the five human SFXN, are available in Reference [8].
SFXNs homologues display a high amino acid identity rate in mouse [1], xenopus [5] and human [8]. In humans, SFXN1 and SFXN3 share 76.56% identical amino acids, whereas there is 56.05% identity between SFXN1 and SFXN2 and only 22.04% between SFXN1 and SFXN4. An alignment of human SFXNs is shown in Figure 1. Identity rates between the different human, Drosophila and yeast sideroflexins proteins are described elsewhere [8][9]. The high degree of homology between SFXNs, especially between SFXN1 and SFXN3 in humans, suggests that sideroflexins ensure redundant functions, as it was proposed for the mitochondrial import of serine that seems to be mediated by SFXN1 [8]. This function will be detailed in the section dedicated to the role of SFXN in regulating mitochondrial metabolism (see Section 3.1). Among the five mammalian SFXNs, SFXN4 is the most divergent member, suggesting that this member does not share the same functions (Figure 1). Indeed, SFXN4 was not able to suppress defects caused by the concomitant loss of SFXN1 and SFXN3 in mammalian cells [8]. Interestingly, till now no study has been done to specifically uncover the Fsf1 function. Due to the high degree of similarity between fungal sideroflexin and SFXN proteins from higher eukaryotes, we think that studies on the functions of Fsf1 will certainly lead to huge advances in the SFXN field and may reveal a general function for this family of proteins.
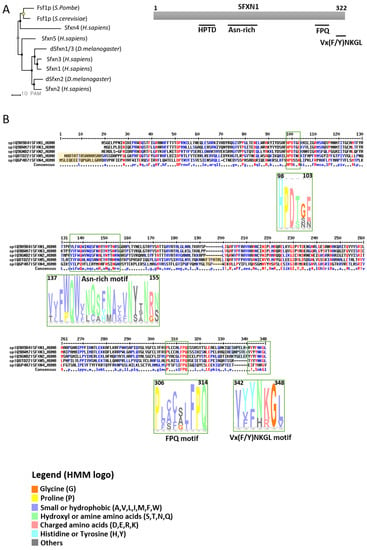
Figure 1. Sideroflexins (SFXNs) form a family of conserved proteins in Eukarya. (A). Left panel: Phylogenetic tree obtained using the MultiAlin software (http://multalin.toulouse.inra.fr/multalin/) [10]. Right panel: scheme of the SFXN1 protein and its conserved motifs. (B). Alignment of human SFXNs protein sequences. Red amino acids are for high consensus levels (90%), the blue ones are for low consensus levels (50%). Meaning of symbols found in the consensus line: “!” is for Ile or Val, “$” is for Leu or Met, “%” is for Phe or Tyr,” #” is anyone of Asn, Asp, Glu, Gln. Conserved motifs are shown and highlighted using an HMM logo created using Skyline (http://skylign.org/) with consensus colors for amino acids according to the ClustalX coloring scheme.
1.3. Sideroflexins Are Mitochondrial Transporters Implicated in One-Carbon Metabolism
SFXNs possess four to six predicted transmembrane domains composed by α-helices revealed by in silico modeling [1][6][7]. These proteins share several highly conserved motifs, including a HPDT motif and an asparagine-rich sequence (Figure 1) [1][6]. The functions of those conserved motifs have not been uncovered yet. Recently, Gyimesi and Hediger performed an in silico analysis of human SFXN1-5 sequences and described six well-conserved regions that could be important for SFXNs activity [11]. Whether these conserved regions are essential for metabolite transport need to be further confirmed at the bench.
To date, no crystal structure has been released for SFXNs. We thus tried to model SFXN tridimensional structure using the trRosetta software [12]. The SFXN1 predicted structure is shown in Figure 2. Interestingly, this structure reveals six internal alpha helices that may correspond to the transmembrane domain of SFXN1.
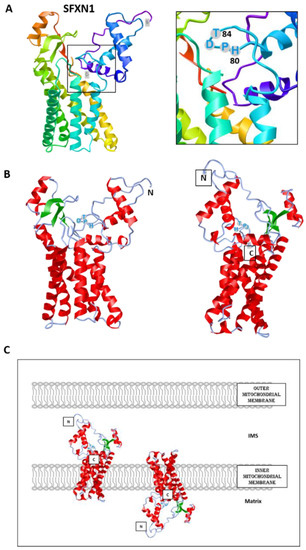
Figure 2. Predicted structure of human SFXN1. Structure prediction was obtained using trRosetta. The confidence of the predicted model shown here is very high (with estimated TM-score = 0.806). The model was built by trRosetta based on de novo folding, guided by deep learning restraints. iCn3D was used for the visualization of 3D structure [13]. (A). SFXN1 predicted structure reveals several alpha helices and beta strands. N and C termini are labelled. The inlet shows the position of the HPDT motif (aa 80–83), located just after the fourth helix. (B). Two views highlighting secondary structures (helices in red, beta sheets in green). (C). Models for SFXN1 insertion in the inner mitochondrial membrane.
SFXN1 topology was recently investigated by APEX and classical biochemical experiments [14][15][16]. Acoba et al. [16] performed detergent extraction and protease-protection assays on HEK human cells and confirmed that endogenous SFXN1 is a mitochondrial protein inserted in the inner mitochondrial membrane (IMM). Furthermore, evidence was given for the presence of the N-terminus in the intermembrane space (IMS), but not in the matrix, contrary to what is predicted by a in silico analysis using Protter. According to biochemical data, the C-terminus seems to protrude in the matrix, in agreement with the previously proposed five transmembrane domains. However, our model is rather in agreement with a TM domain composed of six alpha helices, and if this predicted structure is correct, the N and C termini could be in the same mitochondrial compartment (Figure 2). The CryoEM structure of SFXN1 is thus needed to precise the three-dimensional structure of this carrier. Moreover, two recent studies investigated the mechanisms of SFXN1 mitochondrial import and shed light on the role of TIM22 and AGK2 in this process [16][17]. Evidence for a mitochondrial localization of SFXN is listed in Table 1.
Table 1. Evidence for a mitochondrial localization of Sideroflexins.
| SFXN | Model | Localization | Experiment | Reference |
|---|---|---|---|---|
| SFXN1 | Mouse | IMM | Co-fractionation | Fleming et al. 2001 [1] |
| Human cells (Jurkat, K562) |
Immunoblot on affinity-purified mitochondria STED (co-localization of Flag-SFXN1 and COX4) |
Kory et al. 2018 [8] | ||
| Human cells (MCF7, HT1080), Drosophila | Immunoblot on mitochondrial extracts (fractionation) Confocal microscopy, Proteomics (LC-MS/MS on SFXN1 IP) |
Our unpublished data | ||
| Human cells (HEK) | SILAC-based proteomics coupled LC-MS/MS, carbonate extraction, digitonin fractionation | Acoba et al. 2020 [16] | ||
| SFXN2 | Human cells (HeLa) | OMM or IMM | Confocal microscopy (Tom20 co-localization) | Mon et al. 2018 [9] |
| Human cells (Jurkat, K562) |
Immunoblot on affinity-purified mitochondria | Kory et al. 2018 [8] | ||
| Human cells (HEK) | SILAC-based proteomics coupled LC-MS/MS | Acoba et al. 2020 [16] | ||
| SFXN3 | Rat embryonic brain cells | IMM | Fractionation, Confocal microscopy (co-localization with COX4), TEM | Rivell et al. 2019 [18] |
| Human cells (Jurkat, K562) | Immunoblot on affinity-purified mitochondria | Kory et al. 2018 [8] | ||
| Human cells (HEK) | SILAC-based proteomics coupled LC-MS/MS | Acoba et al. 2020 [16] | ||
| SFXN4 | Human cells (HeLa) | IMM | Fractionation and protease protection assay | Hildick-Smith et al. 2013 [19] |
| Human cells (Jurkat, K562) | Immunoblot on affinity-purified mitochondria | Kory et al. 2018 [8] | ||
| Human cells (HEK) | SILAC-based proteomics coupled LC-MS/MS | Acoba et al. 2020 [16] | ||
| SFXN5 | Human cells (HEK) Mouse astrocytes, human cortex and spinal cord |
SILAC-based proteomics coupled LC-MS/MS Immunocapture of GFP-OMM-tagged mitochondria (MitoTag mice), immunostaining |
Acoba et al. 2020 [16] Fecher et al. 2019 [20] |
IMM: inner mitochondrial membrane, IP: immunoprecipitation, OMM: outer mitochondrial membrane, STED: stimulated emission depletion, TEM: Transmission Electron Microscopy, SILAC: Stable isotope labelling of amino acids, LC-MS/MS: Liquid chromatography and tandem mass spectrometry.
Due to their predicted structure, showing several hydrophobic alpha helices, and their mitochondrial location, sideroflexins were proposed to be mitochondrial metabolite transporters. Rat Sfxn3 was presumed to be a tricarboxylate carrier (TCC), and later, Sfxn5 (also known as BBG-TCC) was reported to transport citrate in vitro [21][22]. However, it was only recently that a function of mitochondrial serine transporter was reported for SFXN1 [8].
By a bioinformatic analysis, the S. cerevisiae Fsf1 (YOR271cp) was proposed to be a candidate alpha-isopropylmalate transporter but no experimental data ascertained this function [23]. Similarly, the predicted Fsf1 protein from Schizosaccharomyces pombe, Spac17g6.15c, is annotated as a serine transporter in the database Pombase (https://www.pombase.org/) based on its homology with human SFXN1 [24][25], although it has not been extensively studied.
Since mice lacking Sfxn1 present similar features to that observed in human syndromes caused by a lack of pyridoxine or ALAS2 mutation (X-linked sideroblastic anemia), it was also proposed that Sfxn1 transports pyridoxine (B6 vitamin) inside the mitochondria [1][26]. Since pyridoxine is the precursor of pyridoxal phosphate that serves as a cofactor for ALAS2 (the erythroid specific enzyme catalyzing the first step of heme biosynthesis), SFXN1 could thus directly regulate heme biosynthesis. However, it has been recently reported that human SFXN1 is not able to transport pyridoxine in vitro [8]. Even if we cannot exclude that SFXN1 functions in a complex that is not fully reconstituted in in vitro assays, SFXN1 may not be the carrier for pyridoxine. Mtm1p, SLC25A39 yeast homologue, was suggested to import pyridoxal 5′-phosphate inside the mitochondria [27][28]. However, the substrate specificity of the SLC25A39 carrier remains unknown [29].
Thus, the main role of SFXN1 seems to be the mitochondrial serine import. Inside the mitochondrion, serine can be catabolized by the serine hydroxymethyl transferase (SHMT2) into glycine, an amino acid necessary for ALA synthesis. Thus, the lack of SFXN1 would lead to decreased mitochondrial levels of serine and glycine, leading to ALA synthesis impairment.
1.4. Sideroflexins in Disease
Hildick-Smith et al. described, for the first time, a human syndrome (combined oxidative phosphorylation deficiency-18, OMIM entry # 615578), which was directly associated with the lack of a member of the SFXN family, namely SFXN4 [19]. Patients showed macrocytic anemia and mitochondriopathy non-explainable by other causes, but the lack of SFXN4. Recently, a third patient with SFXN4 mutations was described by Sofou et al. [30]. The three patients with SFXN4 mutations presented with intrauterine growth retardation, mild to severe intellectual disabilities, microcephaly, neonatal lactic acidosis, macrocytic anemia and severe visual impairment. Sofou et al. reported optic nerve hypoplasia in the third case. More recently, some of the mechanisms that could explain those effects in humans were reported in the K562 erythroleukemic cell line [31]. Interestingly, SFXN4 loss-of-function leads to a general decrease in the levels of the respiratory chain complexes I-IV, which could be explained by an impaired Fe-S cluster synthesis, as evidenced by a Fe-S fluorescence assay (FeSFA). Nevertheless, Sofou et al. showed that the effect of SFXN4 decrease would be exclusively in Complex I, but not in the rest of the respiratory chain complexes after muscle biopsy [30]. Despite these discrepancies, which could be due to the different nature of the mutations analyzed in each case, it seems clear that Complex I activity is affected in both studies, which reinforces the hypothesis that SFXN4 could have a role, either direct or indirect, on Fe-S biosynthesis.
Besides the description of mutations in the SFXN4 human gene causing the COXPD18 syndrome, SFXN4 was also reported to be a predisposition gene for familial colorectal cancer (CRC). Hence, rare SFXN4 truncating variants were identified in 3/96 CRC familial cases [32]. An aberrant expression of SFXN1 and SFXN5 was also reported in patients with breast cancer or gliomas [33][34].
2. Sideroflexins and Mitochondrial Respiration
2.1. Overview of the Mitochondrial Respiratory Complexes and the Place of Iron in RC
Oxidative Phosphorylation (OXPHOS) couples the transport of electrons (through a series of mitochondrial respiratory complexes containing redox-active prosthetic groups) to the production of ATP by the mitochondrial ATP synthase, commonly referred to as the complex V of the respiratory chain (Figure 3). Respiratory complexes (RC) are arranged in supercomplexes (SC) and megacomplexes in the inner mitochondrial membrane [35][36]. The Electron Transport Chain (ETC) comprises four RCs (Complex I-IV) containing more than 70 nuclear DNA encoded subunits and 13 mitochondrial DNA (mtDNA) encoded subunits, some of which include iron-sulfur clusters (ISCs) or heme; those iron-containing groups are essential cofactors for electron transport from one complex to another [37][38]. The purpose of this review is not to give an extensive overview of the abundant literature on RC, so we invite the reader to refer to recent reviews for details on the composition, structure and biogenesis of RC [35][38][39].
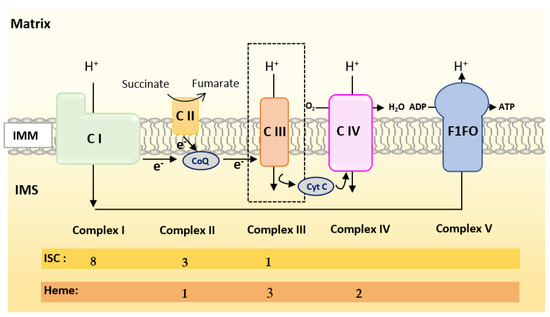
Figure 3. Scheme of the mitochondrial respiratory chain. For each complex, Iron Sulfur Cluster (ISC) and heme numbers are given.
Mammalian Complex I (NADH: Ubiquinone Oxireductase) is a L-shaped megastructure of about 1 MDa comprising 14 core subunits and up to 45 subunits. Among them, five essential subunits (NDUFV1, NUDFV2, NDUFS1, NDUFS7 and NDUFS8) bare the eight ISCs of CI (two [2Fe-2S] and six [4Fe-4S] clusters).
Mammalian Complex II, the smallest of the RC, is composed of only four subunits: succinate dehydrogenase [ubiquinone] flavoprotein (also known as Flavoprotein subunit of complex II, Fp, SDHA), succinate dehydrogenase [ubiquinone] iron-sulfur subunit (a Fe-S protein also named Ip or SDHB), the membrane-anchoring succinate dehydrogenase cytochrome b560 subunit (CybL, SDHC), and finally the succinate dehydrogenase (ubiquinone) cytochrome b small subunit (CybS, SDHD). These subunits are, respectively, encoded by the SDHA, SDHB, SDHC and SDHD nuclear genes. Fp/SDHA and Ip/SDHB are anchored to the IMM thanks to CybL/SDHC and CyBS/SDHD that are the membrane-anchoring subunits of CII. Complex II contains three ICSs ([2Fe-2S], [4Fe-4S] and [3Fe-4S] in SDHB) and a heme shared by SDHC and SDHD.
Mammalian Complex III (also known as bc1 complex) is a dimer made of monomers containing 11 subunits among which three are essential redox subunits: cytochrome b, cytochrome c1 and the Fe-S protein Cytochrome b-c1 complex subunit Rieske (Rieske, ISP, RISP, Rip1 are alternative names that can be found in the literature for this protein). Altogether, these catalytic subunits possess two heme b (Cyt b), a c-type heme (Cyt c1) and a [2Fe-2S] cluster (Rieske) [40]. Heme b is synthesized by Ferrochelatase (FECH), but the mechanism of its insertion into cytochrome b has not been fully elucidated [40].
Mammalian Complex IV contains three mitochondrially-encoded subunits (Cytochrome c oxidase subunit 1, 2 and 3) plus eleven subunits encoded by the nuclear genome. CIV possesses four redox-active metal centers, including heme a and heme a3, but no ISCs.
To summarize, Complex I is made of numerous subunits including 8 ISC-containing subunits but none containing heme. Complex IV presents four redox-active centers containing heme, but no ISC. Both Complexes II and III have ISC and heme containing subunits.
2.2. Current Knowledge on the Regulation of Mitochondrial Respiration by SFXN Proteins
Kory et al. reported decreased basal respiration in SFXN1/SFXN3 double knockout Jurkat cells [8]. Whereas SFXN1 loss alone is not detrimental for respiration of intact cells [8][16], Acoba et al. reported a significant decrease in Oxygen Consumption Rates (OCR) of isolated mitochondria from HEK SFXN1 KO cells with CI, CII and CIII substrates (pyruvate, Glu, Gln, dimethyl-α-ketoglutarate, succinate and glycerol-3-phosphate) [16]. In human embryonic cells, the loss of SFXN1 leads to a marked decrease in the protein levels of three subunits of the Complex III and to a lesser extent in Complex II subunit SDHB (Table 2) [16]. SFXN4 KO leukemic cells also showed reduced levels of several RC subunits containing ISCs [31].
Table 2. Consequences of sideroflexins (SFXN) deficiency on the electron transport chain (ETC) complexes.
| SFXN | Model | Complex | Data | Reference |
|---|---|---|---|---|
| SFXN1 | HEK SFXN1 KO cells HeLa SFXN1 KO cells |
CI | No significant loss of activity SDHB ↓ |
Acoba et al. 2020 [16] |
| CII | No significant loss of activity UQCRC2 ↓↓ UQCRFS1 ↓↓ |
|||
| CIII | Cytochrome b ↓↓↓ Significant loss of activity Reduced levels of CIII2 and CIII2-CIV respiratory complexes |
|||
| SFXN2 | HEK SFXN2 KO cells | CI CII-CIII CIV |
No significant loss of activity Significant loss of activity Significant loss of activity |
Mon et al. 2019 [9] |
| SFXN3 | SFXN3 KO mouse | CI, CIV | No significant loss of activity | Amorim et al. 2017 [45] |
| SFXN4 | Primary fibroblasts from two individuals with SFXN4 mutations | CI + CIII | Decreased activity | Hildick-Smith et al. 2013 [19] |
| SFXN4 KD zebrafish | CI CI + CIII |
Decreased activity | Sofou et al. 2019 [30] | |
| K562 SFXN4 KO cells | CI CII CIII CIV |
NDUFB8 ↓ SDHB ↓ UQCRC2 ↓ COX2 ↓ |
Paul et al. 2019 [31] | |
| SFXN5 | N.A. 1 |
↓ indicates decreased levels of respiratory complexes subunits (↓ low, ↓↓ medium, ↓↓↓ high); 1 N.A.: Not addressed.
Whereas no significant change in the activity of the CI, CII and CIV ETC complexes was observed upon SFXN1 gene knockout in HEK cells, CIII activity was dramatically decreased and partially restored upon SFXN1 overexpression [16]. In agreement with the observed decrease in the levels of cytochrome b (MT-CYB), cytochrome b-c1 complex subunit 2 (UQCRC2) and cytochrome b-c1 complex subunit Rieske (UQCRFS1) subunits, Acoba et al. also reported a reduction in CIII2 and in CIII2-CIV subcomplex, whereas the assembly of respiratory supercomplexes was unaffected. Mitochondrial translation is not dramatically impaired in the absence of a functional SFXN1 protein, nevertheless a slight decrease in cytochrome b translation was reported in this study.
No decrease in either the quantity of mtDNA or in the mitochondrial mass was seen in SFXN1 KO cells; thus, a general defect in mitochondrial biogenesis can be excluded [8][16]. Current knowledge on Complex III biogenesis is well-described in Reference [40]. Seven assembly factors are implicated in CIII biogenesis in humans (UQCC1-3, CCHL, BCSL1, LYRM7 and TTC19). The Rieske subunit is first translocated from the cytosol to the matrix where it acquires its ISC and is further incorporated in CIII. In the matrix, Rieske is stabilized by the chaperone LYRM7 [41]. BCS1L is required for the translocation of the folded Rieske iron-sulfur protein in the IMM by a mechanism that remains largely unknown [42]. No regulation of the levels of BCSL1 and LYRM7 assembly factors was observed when SFXN1 is absent in mammalian cells [16].
Interestingly, HEK SFXN1 KO cells were reported to have markedly reduced levels of Coenzyme Q (CoQ, ubiquinone), a lipid of the IMM that accepts electron from CI and CII and then donates one electron to the ISC of the Rieske subunit and another one to the heme of the cytochrome b of CIII (see Reference [40] and [43] for more details on the transfer of electrons from CoQ to the IMS soluble electron carrier cytochrome c).
Deficiencies of mitochondrial respiration and/or RC activity were also reported for other SFXN, as summarized in Table 2. For example, SFXN2 knockout led to a decreased activity of CII-CIII and CIV [9]. As no specific impairment in complex III activity has been described nor in SFXN2 nor in SFXN4 KO cells, there is presumably no interaction between those SFXN isoforms and the BCS1L protein (responsible of the GRACILE Syndrome), a mitochondrial chaperone that is anchored to the inner mitochondrial membrane and required for proper Complex III activity [44]. Nevertheless, this possibility cannot be totally discarded, as the patients with S78G point mutation in the BCS1L gene have no decreased Complex III activity when compared to other mutations of the same gene.
This entry is adapted from the peer-reviewed paper 10.3390/biomedicines9020103
References
- Fleming, M.D. A Mutation in a Mitochondrial Transmembrane Protein Is Responsible for the Pleiotropic Hematological and Skeletal Phenotype of Flexed-Tail (f/f) Mice. Genes Dev. 2001, 15, 652–657, doi:10.1101/gad.873001.
- Lenox, L.E.; Perry, J.M.; Paulson, R.F. BMP4 and Madh5 Regulate the Erythroid Response to Acute Anemia. Blood 2005, 105, 2741–2748, doi:10.1182/blood-2004-02-0703.
- Hegde, S.; Lenox, L.E.; Lariviere, A.; Porayette, P.; Perry, J.M.; Yon, M.; Paulson, R.F. An Intronic Sequence Mutated in Flexed-Tail Mice Regulates Splicing of Smad5. Mamm Genome 2007, 18, 852–860, doi:10.1007/s00335-007-9074-9.
- SLC56 Sideroflexins. IUPHAR/BPS Guide to PHARMACOLOGY. 2020. Available online: http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=1008 (Accessed on 20 January 2021).
- Li, X.; Han, D.; Kin Ting Kam, R.; Guo, X.; Chen, M.; Yang, Y.; Zhao, H.; Chen, Y. Developmental Expression of Sideroflexin Family Genes in Xenopus Embryos. Dev. Dyn. 2010, 239, 2742–2747, doi:10.1002/dvdy.22401.
- Lockhart, P.J.; Holtom, B.; Lincoln, S.; Hussey, J.; Zimprich, A.; Gasser, T.; Wszolek, Z.K.; Hardy, J.; Farrer, M.J. The Human Sideroflexin 5 (SFXN5) Gene: Sequence, Expression Analysis and Exclusion as a Candidate for PARK3. Gene 2002, 285, 229–237.
- Miotto, G.; Tessaro, S.; Rotta, G.A.; Bonatto, D. In Silico Analyses of Fsf1 Sequences, a New Group of Fungal Proteins Orthologous to the Metazoan Sideroblastic Anemia-Related Sideroflexin Family. Fungal Genet. Biol. 2007, 44, 740–753, doi:10.1016/j.fgb.2006.12.004.
- Kory, N.; Wyant, G.A.; Prakash, G.; Uit de Bos, J.; Bottanelli, F.; Pacold, M.E.; Chan, S.H.; Lewis, C.A.; Wang, T.; Keys, H.R.; et al. SFXN1 Is a Mitochondrial Serine Transporter Required for One-Carbon Metabolism. Science 2018, 362, eaat9528, doi:10.1126/science.aat9528.
- Mon, E.E.; Wei, F.-Y.; Ahmad, R.N.R.; Yamamoto, T.; Moroishi, T.; Tomizawa, K. Regulation of Mitochondrial Iron Homeostasis by Sideroflexin 2. J. Physiol. Sci. 2019, 69, 359–373, doi:10.1007/s12576-018-0652-2.
- Corpet, F. Multiple Sequence Alignment with Hierarchical Clustering. Nucleic Acids Res. 1988, 16, 10881–10890, doi:10.1093/nar/16.22.10881.
- Gyimesi, G.; Hediger, M.A. Sequence Features of Mitochondrial Transporter Protein Families. Biomolecules 2020, 10, 1611, doi:10.3390/biom10121611.
- Yang, J.; Anishchenko, I.; Park, H.; Peng, Z.; Ovchinnikov, S.; Baker, D. Improved Protein Structure Prediction Using Predicted Interresidue Orientations. Proc. Natl. Acad. Sci. USA 2020, 117, 1496–1503, doi:10.1073/pnas.1914677117.
- Wang, J.; Youkharibache, P.; Zhang, D.; Lanczycki, C.J.; Geer, R.C.; Madej, T.; Phan, L.; Ward, M.; Lu, S.; Marchler, G.H.; et al. ICn3D, a Web-Based 3D Viewer for Sharing 1D/2D/3D Representations of Biomolecular Structures. Bioinformatics 2020, 36, 131–135, doi:10.1093/bioinformatics/btz502.
- Yoo, C.-M.; Rhee, H.-W. APEX, a Master Key to Resolve Membrane Topology in Live Cells. Biochemistry 2019, doi:10.1021/acs.biochem.9b00785.
- Lee, S.-Y.; Kang, M.-G.; Park, J.-S.; Lee, G.; Ting, A.Y.; Rhee, H.-W. APEX Fingerprinting Reveals the Subcellular Localization of Proteins of Interest. Cell Rep. 2016, 15, 1837–1847, doi:10.1016/j.celrep.2016.04.064.
- Acoba, M.G.; Selen Alpergin, E.S.; Renuse, S.; Fernández-del-Río, L.; Lu, Y.-W.; Clarke, C.F.; Pandey, A.; Wolfgang, M.J.; Claypool, S.M. The Mitochondrial Carrier SFXN1 Is Critical for Complex III Integrity and Cellular Metabolism; Biochemistry; 2020. doi: https://doi.org/10.1101/2020.06.18.157495
- Jackson, T.D.; Hock, D.; Palmer, C.S.; Kang, Y.; Fujihara, K.M.; Clemons, N.J.; Thorburn, D.R.; Stroud, D.A.; Stojanovski, D. The TIM22 Complex Regulates Mitochondrial One-Carbon Metabolism by Mediating the Import of Sideroflexins; Cell Biology; 2020. doi: https://doi.org/10.1101/2020.02.06.937920.
- Rivell, A.; Petralia, R.S.; Wang, Y.-X.; Mattson, M.P.; Yao, P.J. Sideroflexin 3 Is a Mitochondrial Protein Enriched in Neurons. Neuromol. Med. 2019, 21, 314–321, doi:10.1007/s12017-019-08553-7.
- Hildick-Smith, G.J.; Cooney, J.D.; Garone, C.; Kremer, L.S.; Haack, T.B.; Thon, J.N.; Miyata, N.; Lieber, D.S.; Calvo, S.E.; Akman, H.O.; et al. Macrocytic Anemia and Mitochondriopathy Resulting from a Defect in Sideroflexin 4. Am. J. Hum. Genet. 2013, 93, 906–914, doi:10.1016/j.ajhg.2013.09.011.
- Fecher, C.; Trovò, L.; Müller, S.A.; Snaidero, N.; Wettmarshausen, J.; Heink, S.; Ortiz, O.; Wagner, I.; Kühn, R.; Hartmann, J.; et al. Cell-Type-Specific Profiling of Brain Mitochondria Reveals Functional and Molecular Diversity. Nat. Neurosci. 2019, 22, 1731–1742, doi:10.1038/s41593-019-0479-z.
- Azzi, A.; Glerum, M.; Koller, R.; Mertens, W.; Spycher, S. The Mitochondrial Tricarboxylate Carrier. J. Bioenerg. Biomembr. 1993, 25, 515–524, doi:10.1007/BF01108408.
- Miyake, S.; Yamashita, T.; Taniguchi, M.; Tamatani, M.; Sato, K.; Tohyama, M. Identification and Characterization of a Novel Mitochondrial Tricarboxylate Carrier. Biochem. Biophys. Res. Commun. 2002, 295, 463–468, doi:10.1016/S0006-291X(02)00694-0.
- Kovaleva, G.Yu.; Bazykin, G.A.; Brudno, M.; Gelfand, M.S. COMPARATIVE GENOMICS OF TRANSCRIPTIONAL REGULATION IN YEASTS AND ITS APPLICATION TO IDENTIFICATION OF A CANDIDATE ALPHA-ISOPROPYLMALATE TRANSPORTER. J. Bioinform. Comput. Biol. 2006, 04, 981–998, doi:10.1142/S0219720006002284.
- Wood, V.; Harris, M.A.; McDowall, M.D.; Rutherford, K.; Vaughan, B.W.; Staines, D.M.; Aslett, M.; Lock, A.; Bahler, J.; Kersey, P.J.; et al. PomBase: A Comprehensive Online Resource for Fission Yeast. Nucleic Acids Res. 2012, 40, D695–D699, doi:10.1093/nar/gkr853.
- Lock, A.; Rutherford, K.; Harris, M.A.; Hayles, J.; Oliver, S.G.; Bähler, J.; Wood, V. PomBase 2018: User-Driven Reimplementation of the Fission Yeast Database Provides Rapid and Intuitive Access to Diverse, Interconnected Information. Nucleic Acids Res. 2019, 47, D821–D827, doi:10.1093/nar/gky961.
- Ye, X.; Xu, J.; Cheng, C.; Yin, G.; Zeng, L.; Ji, C.; Gu, S.; Xie, Y.; Mao, Y. Isolation and Characterization of a Novel Human Putative Anemia-Related Gene Homologous to Mouse Sideroflexin. Biochem. Genet. 2003, 41, 119–125.
- Whittaker, M.M.; Penmatsa, A.; Whittaker, J.W. The Mtm1p Carrier and Pyridoxal 5′-Phosphate Cofactor Trafficking in Yeast Mitochondria. Arch. Biochem. Biophys. 2015, 568, 64–70, doi:10.1016/j.abb.2015.01.021.
- Whittaker, J.W. Intracellular Trafficking of the Pyridoxal Cofactor. Implications for Health and Metabolic Disease. Arch. Biochem. Biophys. 2016, 592, 20–26, doi:10.1016/j.abb.2015.11.031.
- Curcio, R.; Lunetti, P.; Zara, V.; Ferramosca, A.; Marra, F.; Fiermonte, G.; Cappello, A.R.; De Leonardis, F.; Capobianco, L.; Dolce, V. Drosophila Melanogaster Mitochondrial Carriers: Similarities and Differences with the Human Carriers. IJMS 2020, 21, 6052, doi:10.3390/ijms21176052.
- Sofou, K.; Hedberg-Oldfors, C.; Kollberg, G.; Thomsen, C.; Wiksell, Å.; Oldfors, A.; Tulinius, M. Prenatal onset of mitochondrial disease is associated with sideroflexin 4 deficiency. Mitochondrion 2019, 47, 76–81, doi:10.1016/j.mito.2019.04.012.
- Paul, B.T.; Tesfay, L.; Winkler, C.R.; Torti, F.M.; Torti, S.V. Sideroflexin 4 Affects Fe-S Cluster Biogenesis, Iron Metabolism, Mitochondrial Respiration and Heme Biosynthetic Enzymes. Sci. Rep. 2019, 9, 19634, doi:10.1038/s41598-019-55907-z.
- Gylfe, A.E.; Katainen, R.; Kondelin, J.; Tanskanen, T.; Cajuso, T.; Hänninen, U.; Taipale, J.; Taipale, M.; Renkonen-Sinisalo, L.; Järvinen, H.; et al. Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer. PLOS Genet 2013, 9, doi:10.1371/journal.pgen.1003876.
- Weston, C.; Klobusicky, J.; Weston, J.; Connor, J.; Toms, S.A.; Marko, N.F. Aberrations in the Iron Regulatory Gene Signature Are Associated with Decreased Survival in Diffuse Infiltrating Gliomas. PLoS ONE 2016, 11, e0166593, doi:10.1371/journal.pone.0166593.
- Miller, L.D.; Coffman, L.G.; Chou, J.W.; Black, M.A.; Bergh, J.; D’Agostino, R.; Torti, S.V.; Torti, F.M. An Iron Regulatory Gene Signature Predicts Outcome in Breast Cancer. Cancer Res. 2011, 71, 6728–6737, doi:10.1158/0008-5472.CAN-11-1870.
- Wu, M.; Gu, J.; Zong, S.; Guo, R.; Liu, T.; Yang, M. Research Journey of Respirasome. Protein Cell 2020, 11, 318–338, doi:10.1007/s13238-019-00681-x.
- Guo, R.; Gu, J.; Zong, S.; Wu, M.; Yang, M. Structure and Mechanism of Mitochondrial Electron Transport Chain. Biomed. J. 2018, 41, 9–20, doi:10.1016/j.bj.2017.12.001.
- Stiban, J.; So, M.; Kaguni, L.S. Iron-Sulfur Clusters in Mitochondrial Metabolism: Multifaceted Roles of a Simple Cofactor. Biochem. Mosc. 2016, 81, 1066–1080, doi:10.1134/S0006297916100059.
- Barros, M.H.; McStay, G.P. Modular Biogenesis of Mitochondrial Respiratory Complexes. Mitochondrion 2020, 50, 94–114, doi:10.1016/j.mito.2019.10.008.
- Letts, J.A.; Sazanov, L.A. Clarifying the Supercomplex: The Higher-Order Organization of the Mitochondrial Electron Transport Chain. Nat. Struct. Mol. Biol. 2017, 24, 800–808, doi:10.1038/nsmb.3460.
- Ndi, M.; Marin-Buera, L.; Salvatori, R.; Singh, A.P.; Ott, M. Biogenesis of the Bc1 Complex of the Mitochondrial Respiratory Chain. J. Mol. Biol. 2018, 430, 3892–3905, doi:10.1016/j.jmb.2018.04.036.
- Sánchez, E.; Lobo, T.; Fox, J.L.; Zeviani, M.; Winge, D.R.; Fernández-Vizarra, E. LYRM7/MZM1L Is a UQCRFS1 Chaperone Involved in the Last Steps of Mitochondrial Complex III Assembly in Human Cells. Biochim. et Biophys. Acta (BBA) Bioenerg. 2013, 1827, 285–293, doi:10.1016/j.bbabio.2012.11.003.
- Tang, W.K.; Borgnia, M.J.; Hsu, A.L.; Esser, L.; Fox, T.; de Val, N.; Xia, D. Structures of AAA Protein Translocase Bcs1 Suggest Translocation Mechanism of a Folded Protein. Nat. Struct. Mol .Biol. 2020, 27, 202–209, doi:10.1038/s41594-020-0373-0.
- Wang, Y.; Hekimi, S. Understanding Ubiquinone. Trends Cell Biol. 2016, 26, 367–378, doi:10.1016/j.tcb.2015.12.007.
- Visapää, I.; Fellman, V.; Vesa, J.; Dasvarma, A.; Hutton, J.L.; Kumar, V.; Payne, G.S.; Makarow, M.; Van Coster, R.; Taylor, R.W.; et al. GRACILE Syndrome, a Lethal Metabolic Disorder with Iron Overload, Is Caused by a Point Mutation in BCS1L. Am. J. Hum. Genet. 2002, 71, 863–876, doi:10.1086/342773.
- Amorim, I.S.; Graham, L.C.; Carter, R.N.; Morton, N.M.; Hammachi, F.; Kunath, T.; Pennetta, G.; Carpanini, S.M.; Manson, J.C.; Lamont, D.J.; et al. Sideroflexin 3 Is an α-Synuclein-Dependent Mitochondrial Protein That Regulates Synaptic Morphology. J. Cell. Sci. 2017, 130, 325–331, doi:10.1242/jcs.194241.
