Emerging infectious diseases present great risks to public health. The novel severe acute respira-tory syndrome coronavirus 2 (SARS-CoV-2), causing coronavirus disease 2019 (COVID-19), has become an urgent public health issue of global concern. It is speculated that the virus first emerged through a zoonotic spillover. Basic research studies have suggested that bats are likely the ancestral reservoir host. Nonetheless, the evolutionary history and host susceptibility of SARS-CoV-2 remains unclear as a multitude of animals has been proposed as potential intermedi-ate or dead-end hosts. SARS-CoV-2 has been isolated from domestic animals, both companion and livestock, as well as in captive wildlife that were in close contact with human COVID-19 cases. Currently, domestic mink is the only known animal that is susceptible to a natural infection, develop severe illness, and can also transmit SARS-CoV-2 to other minks and humans.
- coronavirus
- SARS-CoV-2
- host diversity
- One Health
- COVID-19
- animals
- humans
Note: The following contents are extract from your paper. The entry will be online only after author check and submit it.
1. Introduction
Coronaviruses (CoVs) (order: Nidovirales, family: Coronaviridae, subfamily: Coronavirinae) are enveloped, positive-stranded RNA viruses [1–4]. CoVs can infect birds (Gammacoronaviruses and Deltacoronaviruses) or mammals (predominantly Alpacoronaviruses and Betacoronaviruses) [5,6]. For over 80 years, animal coronaviruses, such as transmissible gastroenteritis virus (TGEV) of swine or bovine CoV (BCoV), have been known to infect wildlife and livestock species [7]. To date, seven CoVs have been identified in humans: HCoV-OC53, HCoV-229E, HCoV-NL63, HCoV-HKU1, MERS-CoV, SARS-CoV, and SARS-CoV-2. The first reports of endemic human CoVs (HCoVs) were documented in the 1960s when HCoV-OC53 and HCoV-229E were described [8,9]. It was not until 2004 and 2005 that HCoV-NL63 and HCoV-HKU1 were detected, respectively [10,11]. Endemic human coronaviruses most likely evolved from ancestral viruses of animal reservoirs [6,12].
In 2003, severe acute respiratory syndrome coronavirus (SARS-CoV) was reported as the first CoV of global health importance, which originated from several horseshoe bat species before transmission into human populations [13]. At least 8000 infections and 774 mortalities were linked to SARS-CoV [14]. Less than a decade later, the Middle Eastern respiratory syndrome (MERS) illness caused by a coronavirus (MERS-CoV) became an endemic disease throughout the Middle East, Africa, and Southeast Asia [15]. The zoonotic origins of MERS-CoV remain unclear, but it is speculated that the virus was transmitted from bat species to dromedary camels in the distant past [15,16].
In December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first detected in the Huanan South seafood market (HSSM), a large market that also traded live animals, within Wuhan City, Hubei province, China [17]. In addition to fish and shellfish, a diverse selection of live wildlife, including hedgehogs, badgers, snakes, and poultry, was marketed at the time when the outbreak occurred [18]. Aside from live wildlife, animal-food products such as carcasses and meat were also available [19]. Here, it is suggested that several clusters of pneumonia cases were linked to the HSSM [20,21]. Phylodynamic analysis reveals that SARS-CoV-2 most likely emerged as early as October 2019 [22,23], suggesting that the HSSM was mainly a super spreading location and not an index spillover event. Shortly after, SARS-CoV-2 spread globally and the World Health Organization (WHO) declared it a pandemic on 11 of March 2020 [24]. As of 4 of January 2021, approximately 1,839,660 million people have died from the novel coronavirus disease 2019 (COVID-19) and more than 83,910,386 million have been infected worldwide [24]. Following 229E-CoV, NL63-CoV, OC43-CoV, HKU1-CoV, SARS-CoV, and MERS-CoV, SARS-CoV-2 is the seventh coronavirus to infect humans.
SARS-CoV and SARS-CoV-2 belong to the subgenus Sarbecoviruses, characterized by frequent recombination events [25,26]. To date, research indicates that SARS-CoV-2 is not an outcome of a recombination event of any known Sarbecoviruses [27]. It is hypothesized that SARS-CoV-2 originated from an unknown animal reservoir [20,21,28–30]. Currently, the closest related sequences originated from horseshoe bat (96%) and pangolin CoVs (91%) [31,32]. Although the receptor-binding domain (RBD) between pangolin CoV is structurally identical to SARS-CoV-2 [33,34], it is unclear if pangolins function as intermediate or dead-end hosts [35–37]. Moreover, a diverse array of mammalian, avian, and reptilian species have been proposed as other potential intermediate hosts [38–41] Narrow genomic variation in CoVs can lead to wide host diversity as demonstrated by the similarity of SARS-CoV-2 to SARS-CoV and MERS-CoV, sharing 99.8% [36] and 99.5% [42] similarity to that from civet cats and dromedary camels, respectively. Consequently, minimal genetic variation is needed for CoVs to exhibit unique host specificity. Therefore, numerous mammalian, avian, and reptilian species have been proposed as potential hosts of SARS-CoV-2 [38,39,41,43–45].
Here, we provide an overview of the host diversity SARS-CoV-2 to veterinary and public health interventions. Evidence in support of reverse zoonotic transmission has been reported in numerous settings where infected humans have engaged in close contact with domestic and captive zoo animals [40,46]. Mink is the only animal to date that has been shown to transmit SARS-CoV-2 to humans, however, we cannot exclude a SARS-CoV-2 transmission potential from cats, dogs, and ferrets to humans. Further studies are needed to elucidate this hypothesis. Moreover, in selected animal groups, there is evidence that animals were infected by SARS-CoV-2 from humans, followed by a subsequent zoonotic transmission of SARS-CoV-2 from these same animals back to human populations [46,47]. This review aims to provide a cross-disciplinary, “One Health” approach to evaluate the SARS-CoV-2 emergence and spread at the intersection of humans and animals [38]. Based on the definition from the Centers for Disease Control and Prevention (CDC): “One Health is a collaborative, multisectoral, and transdisciplinary approach—working at the local, regional, national, and global levels—with the goal of achieving optimal health outcomes recognizing the interconnection between people, animals, plants, and their shared environment” [48]. Furthermore, these findings might support future surveillance programs to unravel the complex evolutionary histories of SARS-CoV-2 and those of SARS-CoV-like CoV viruses of other animal host species.
2. Epidemiology of Human SARS-CoV-2 Infections
Contextual understanding of the epidemiology of the virus is essential to properly study the epidemiology of SARS-CoV-2. Since the initial outbreak in Wuhan, most research on SARS-CoV-2 transmission has been collected through human-to-human transmission studies [49]. Initial studies of SARS-CoV-2 have indicated that the reproductive number (R0) in humans varies from 1.4 to 3.9 [50–54], and approximately 40 to 50% of SARS-CoV-2 human cases are asymptomatic [55–59]. The incubation period for COVID-19 is speculated to be 14 days alongside a median time of 4–5 days from exposure to symptoms onset [60–62]. Global disease trends suggest that women exhibit stronger immune responses than men and they have lower mortality rates [63,64]. Moreover, living at high altitudes has been suggested as a potential natural protective effect for lower mortality [65,66]. Additionally, viral transmission varies by geographic region due to differences in cases’ demographics, genetics, and health behavior practices [36,53,67].
At the population level, systematic health and socioeconomic inequalities have placed many marginalized groups at increased risk of high morbidity and mortality of SARS-CoV-2 infections [68,69]. Previous studies documented that racial and ethnic minorities are disproportionately higher affected by SARS-CoV-2 infections [70,71]. In many of these cases, social determinants have historically limited these groups from accessing fair opportunities for economic, physical, and emotional health [72]. Moreover, socioeconomic status has been linked to the availability of housing and housing conditions (i.e., the number of individuals per household) [73,74]. Living conditions, such as homelessness and crowded living environments (e.g., prisons, nursing homes, and orphanages) have been reported to be associated with increased SARS-CoV-2 infections [75,76].
At the individual level, older adults and people with underlying medical conditions are at higher risk for a severe SARS-CoV-2 illness [77]. In contrast to these groups, most infected children that express symptoms, if any, are generally mild and require only supportive care [56,78]. According to the CDC, some examples of underlying medical and physical conditions that could increase the risk of severe SARS-CoV-2 illness include cancer, chronic kidney disease, heart conditions, obesity (body mass index (BMI) of 30 kg/m2 or higher but <40 kg/m2), severe obesity (BMI ≥ 40 kg/m2), and diabetes mellitus [77]. Other individual-level risk factors include people with disabilities, developmental and behavioral disorders, and drug and substance use disorders [77].
3. Transmission Pathways of SARS-CoV-2 in Humans
Transmission of SARS-CoV-2 among humans is thought to occur via three primary pathways: (1) inhaling respiratory droplets from an infected individual, (2) inhaling infected airborne particles, (3) or contact with infected environmental surfaces also known as fomites [79]. Indirect or direct contact with infected people can facilitate the exposure to infected saliva and other respiratory secretions, commonly excreted when an infected individual coughs, sneezes, talks, or sings [80–83]. It is important to note that the diameter of respiratory droplets (>5–10 μm) is typically larger than that of nuclei or aerosols (>5 μm) [84]. Therefore, the transmission of infected respiratory droplets can occur when a susceptible individual is within 1 m of an infected case [85].
In contrast to droplet transmission, airborne transmission can occur mostly indoors through the dissemination of infectious aerosols that can be suspended in the air for long distances (usually greater than 2 m) and periods (typically hours) [86,87]. Experimental studies that have created infectious aerosols in controlled laboratory settings demonstrated that SARS-CoV-2 can persist in the air from 3 to 16 h [87,88]. Additionally, respiratory excretions from infected individuals can contaminate a variety of surfaces, thus creating fomites that can infect other individuals in the immediate environment upon contact followed by touching the mouth, nose, or eyes [85].
In general, microenvironmental characteristics such as ambient temperature, pH, and humidity greatly impact the persistence of SARS-CoV-2 on surfaces [89]. Similar to other human and animal CoVs [89], SARS-CoV-2 also exhibits low persistence on copper, latex, and other limited porosity surfaces compared to metals, glass, and highly porous fabrics [90,91]. Although SARS-CoV-2 has been reported to survive in environments at 40 °C for up to several hours [92], CoVs survive best at lower environmental temperatures and lower relative humidity [89]. While at room temperature, SARS-CoV-2 is stable at a wide range of pH values (pH 3–10) [93]. Despite evidence of SARS-CoV-2 contamination of surfaces and persistence on various substrates, there is no specific study that directly associates SARS-CoV-2 transmission through fomites [85]. Therefore, it is suggested that fomite transmission has lower importance compared to transmission via inhaling infected respiratory droplets or airborne particles [93,94].
Other modes of SARS-CoV-2 transmission could potentially include fecal-oral, bloodborne, and zoonotic transmission. To date, there have been no published reports indicating SARS-CoV-2 transmission through feces or urine [85]. However, SARS-CoV-2 has been found in the feces of COVID-19 patients [60,95,96], leading to successful cultures of SARS-CoV-2 from stool specimens [97,98]. Additionally, levels of SARS-CoV-2 RNA concentrations in municipal wastewater parallel trends in local COVID-19 outbreaks, supporting an additional methodology for tracking SARS-CoV-2 levels in local human populations [99,100].
Previous studies have detected low concentrations of SARS-CoV-2 in plasma or serum [101]. The potential for bloodborne transmission remains unclear but it is unlikely given the low concentration of viral RNA detected from blood [102,103].
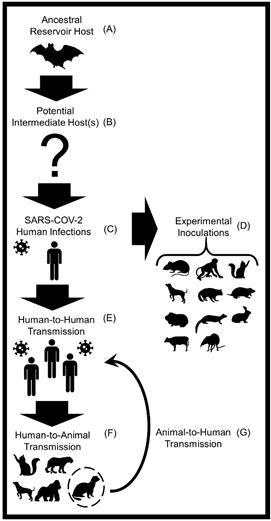
The most recent novel SARS-CoV-2 transmission pathway was described at the human-animal intersection, as current findings suggest a spillback and spillover potential of SARS-CoV-2, especially between humans and domestic mink [46,47] and between humans and companion cats [104]. This synthesis review serves to further evaluate the animal host diversity and zoonotic transmission potential of SARS-CoV-2 (Figure 1).
Figure 1. A conceptual diagram displaying the transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) among humans and various animal hosts. (A) Horseshoe bats (Rhinolophus affinis) are the most likely animal reservoir and ancestral hosts of the SARS-like CoV that gave rise to SARS-CoV-2 [53,105]. (B) A multitude of animals including mammals, birds, and reptiles have been proposed as potential intermediate hosts [38,39,41]. (C) SARS-CoV-2 was first reported in humans in December 2019 in Wuhan, China [106]. (D) Successful laboratory infections of SARS-CoV-2 have been reported in the following mammals: domestic dogs, domestic cats, ferrets, rabbits, raccoon dogs, hamsters, mice, tree shrews, cattle, and several species of non-human primates [107,108]. (E) In January 2020, the World Health Organization (WHO) first reported that human-to-human transmission of SARS-CoV-2 is feasible [109,110]. (F) Natural infections of SARS-CoV-2 in animals transmitted from humans (i.e., reverse zoonosis or anthroponosis) have been detected in domestic dogs and cats, domestic mink, ferrets, mice, hamsters, captive gorillas, and captive large cats (e.g., tigers and lions) [111,112]. (G) Evidence of SARS-CoV-2 spillback from domestic minks to humans and intraspecies transmission of SARS-CoV-2 among minks has been detected [46,47]. At this time these are the described transmission pathways and animals.
This entry is adapted from the peer-reviewed paper 10.3390/pathogens10020180