Comprehensive and concise review on the current knowledge of genetic aberrations in Hodgkin lymphoma.
- Hodgkin lymphoma
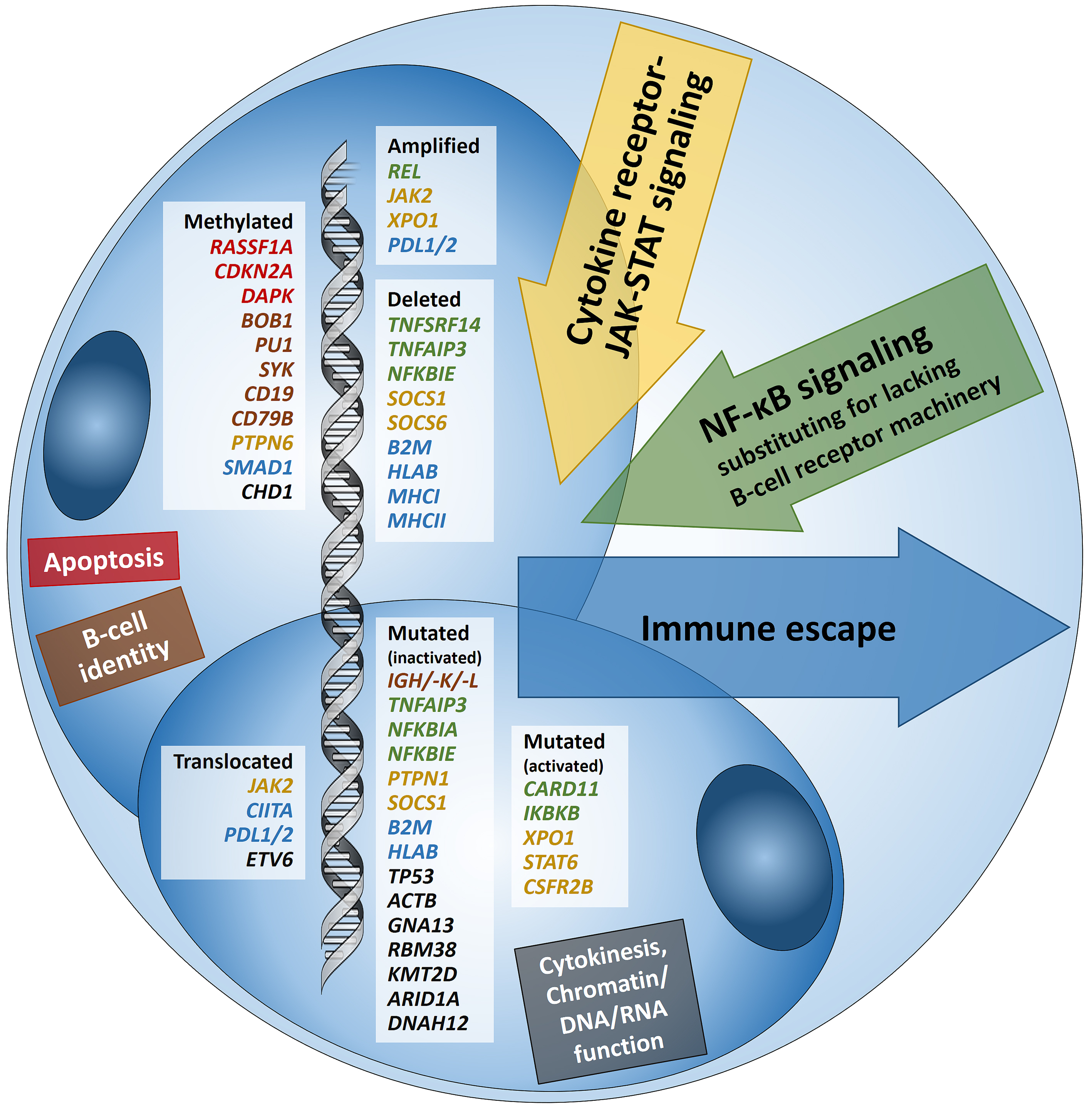
Hodgkin lymphoma (HL) is predominantly composed of reactive, non-neoplastic cells surrounding scarcely distributed tumor cells, that is, so-called Hodgkin and Reed-Sternberg (HRS) or lymphocyte predominant (LP) cells. This scarcity impeded the analysis of the tumor cell genomes for a long time, but recently developed methods (especially laser capture microdissection, flow cytometry/fluorescence-activated cell sorting) facilitated molecular investigation, elucidating the pathophysiological principles of “Hodgkin lymphomagenesis”. The molecular cornerstones of this Hodgkin lymphomagenesis can be summarized as follows (Figure 1): Firstly, the malignant cells of HL evade the immune system by altered expression of PDL1/2, B2M and MHC class I and II due to various genetic alterations. Secondly, tumor growth is promoted by permanently activated JAK/STAT signaling due to pervasive mutations of multiple genes involved in the pathway. Thirdly, apoptosis of neoplastic cells is prevented by alterations of NF-κB compounds and the PI3K/AKT/mTOR axis. Additionally, Epstein-Barr virus infection can simultaneously activate JAK/STAT and NF-κB, similarly leading to enhanced survival and evasion of apoptosis. Finally, epigenetic phenomena such as promoter hypermethylation lead to the downregulation of B-lineage-specific, tumor-suppressor and immune regulation genes.
Figure 1
Figure legend: Summary of known genetic aberrations in classic Hodgkin lymphoma (cHL) arranged according to aberration type and color-coded according to the affected cellular process that they dysregulate in Hodgkin and Reed-Sternberg cells; genes encoding for proteins related to apoptosis are in red, to B-cell identity—in brown, to cytokine (mainly JAK-STAT) signaling—in orange-gold, to NF-κB signaling—in green, to immune escape—in blue, and to cytokinesis, chromatin/DNA/RNA functions—in black.
For more information see: Brune MM, Juskevicius D, Haslbauer J, Dirnhofer S, Tzankov A. Genomic Landscape of Hodgkin Lymphoma. Cancers (Basel). 2021 Feb 8;13(4):682. doi: 10.3390/cancers13040682. PMID: 33567641.
This entry is adapted from the peer-reviewed paper 10.3390/cancers13040682