Liquid–liquid phase separation (LLPS) represents a major physiochemical principle to organize intracellular membrane-less structures. Studies with non-segmented negative-sense (NNS) RNA viruses have uncovered a key role of LLPS in the formation of viral inclusion bodies (IBs), sites of viral protein concentration in the cytoplasm of infected cells. These studies further reveal the structural and functional complexity of viral IB factories as liquid-like organelles and sites of viral replication.
- inclusion body
- viral mediated host remodeling
- liquid–liquid phase separation
- viral replication
- measles virus
- RNA binding protein
- membrane
- biophysical processes
- negative-strand RNA virus
1. Introduction
Among all virus taxa, the phylum Negarnaviricota embodies all viruses with RNA genomes of negative polarity [1]. This includes viruses with either non-segmented negative-sense (NNS) or segmented negative-sense (SNS) RNA genomes. NNS RNA viruses of order Mononegavirales (notably families Paramyxoviridae, Bornaviridae, Rhabdoviridae, Filoviridae, and Pneumoviridae) and SNS RNA viruses such as Influenza A (family Orthomyxoviridae) have historically caused severe human infectious diseases, some with high mortality. These pathogenic viruses have been the center of research efforts to better understand their replication mechanisms at the molecular and cell biological levels [2].
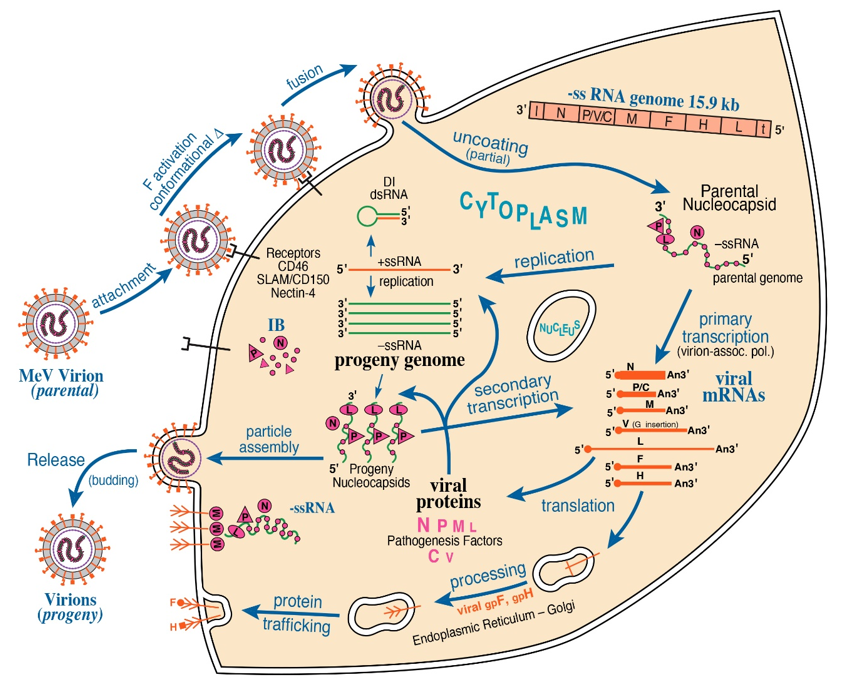
Structure-function studies of NNS and SNS RNA viruses have bridged disciplines between molecular virology and cell biology via elucidating the mechanisms by which viruses replicate. For Negarnaviricota, a virion-associated RNA-dependent RNA Polymerase (vRdRP) is the hallmark of viral mRNA transcription and genome replication [1]. Within the nucleocapsid, both NNS and SNS RNA genomes are encapsidated by viral nucleoprotein (N or NP), forming the stable helical ribonucleoprotein (RNP) complex. Structural studies have been conducted to characterize viral RNPs for several NNS RNA viruses, including vesicular stomatitis virus (VSV) [3], rabies virus (RABV) [4], respiratory syncytial virus (RSV) [5], parainfluenza virus type 5 (PIV5) [6], and Ebola virus (EBOV) [7]. Upon infection by these mononegaviruses, viral macromolecular synthesis is cytoplasmic (with exception of Bornaviridae where it is nuclear). This is illustrated by the Figure 1 schematic summary of measles virus (MeV) replication. The vRdRP Large (L) polymerase protein with polyribonucleotidyltransferase and methyltransferase activities complexes with its viral processivity factor phosphoprotein (P, VP35 for Filoviridae), thereby tethering L to viral RNP for the initiation of de novo RNA synthesis [8][9][10][11]. In contrast, the vRdRP of SNS viruses utilizes a cap-dependent endonuclease to cannibalize host cellular mRNA caps and directly initiates de novo RNA synthesis of each viral RNP segment without the necessity of a phosphoprotein [12][13][14][15]. Collectively, these structural-functional foundations highlight necessary components for viral macromolecular synthesis among the NNS and SNS RNA viruses.
Figure 1. Schematic diagram of measles virus (MeV) multiplication cycle. MeV possesses a negative-sense, single-stranded RNA genome of ~15.9 kb. Virus multiplication occurs in the cytoplasm. Following receptor-mediated virion attachment, the viral envelope fuses with host membranes, releasing the ribonucleocapsid (RNP) complex consisting of genome RNA encapsidated by nucleoprotein N into the cytoplasm; this triggers activation of the virion-associated RNA polymerase complex of viral L and P proteins. The RNP constitutes the basic machinery responsible for RNA synthesis, initially for transcription and then for replication. These processes are thought to be associated with the cytoplasmic inclusion body (IB) factories. Viral C protein interacts with P and enhances polymerase processivity; in the absence of C, defective interfering (DI) RNAs are readily formed. IB assembly triggered by MeV infection is mediated by liquid–liquid phase separation (LLPS). MeV N and P proteins are sufficient to form IB structures by LLPS. The viral matrix M protein mediates assembly of progeny enveloped virions that include two viral glycoproteins, the hemagglutinin H attachment protein and the F fusion protein.
Inspired by theories from soft matter physics, liquid phase condensates are an emerging paradigm for the intracellular organization of membrane-less organelles [16][17][18][19], which differentiates them from classic membrane-bound organelles. Membrane-less and dynamic, these biomolecular condensates compartmentalize diverse structures such as the worm P granules [20], the chromatin [21][22][23], nucleolus [24][25], Cajal bodies [26], PML bodies [27], nuclear speckles [28], P bodies [29], stress granules [30][31], and spindle apparatus [32]. Condensate formation has also been implicated in membrane deformation [33], cargo sorting [34], autophagosome formation [35], transcriptional activation [36][37][38], and stress-induced proteasome-mediated protein degradation in the nucleus [39]. Driven by multivalent macromolecular interactions, these biomolecular condensates are formed via liquid–liquid phase separation (LLPS), with assembly/disassembly regulated by an array of physicochemical factors [16][17][18][19][40]. Notably, biomolecular condensates can adopt a wide range of viscosities: from liquid, to gel, to solid-like states under physiological conditions. For example, unlike the stress granules in human cells that exhibit liquid properties, stress granules in yeast are more solid in nature, presumably the result of adaptation to extreme environments [29]. Similarly, Balbiani bodies of immature oocytes are solid-like structures which likely protect the organelles during oocyte dormancy [41]. On the other hand, the nuclear pore complexes form a gel-like channel which is important for maintaining its permeability barrier [42][43]. Although the functions of these novel phase-separated subcellular structures continue to be elucidated, the primary purposes of these multiphase compartments are thought to increase reaction rates and/or specificities, sequester and/or buffer molecules, and dynamically reorganize molecular hubs [16][17][19].
The authors have observed the aforementioned paradigm of LLPS extending from cell biological contexts to viral molecular condensates. In retrospect, studies on NNS and SNS RNA viruses played a major role by revealing an unappreciated link between LLPS and virology. Here, the authors reviewed studies on viral liquid phase-separated cytoplasmic organelles seen among NNS RNA viruses, exploring their formation, functions, and eventual ageing as viral factories. They discussed future directions of study of these liquid organelles and their prospective roles in the viral replication cycle, host responses to infection, and clinical relevance. The LLPS-mediated cytoplasmic condensate has also been shown for Influenza A virus (IAV, a member of SNS RNA viruses) [44]. Compared to NNS RNA viruses, the influenza viral condensate has a different composition (e.g., the lack of a phosphoprotein) and function (e.g., the lack of a role in genomic replication) [45], and thus was not included in the discussion below.
2. The Nature and Formation of Viral Liquid-Like Organelles
2.1. The Nomenclature
Cytopathic effects from viral infections were well observed in cytological studies and clinical diagnosis [46][47][48], one predominately being the appearance of inclusion bodies (IBs) in the cytoplasm and nucleus. Historically, it has been suggested that either viruses may utilize these inclusions to concentrate viral and host proteins for facilitating viral replication and/or assembly, or the host may orchestrate the inclusion formation in response to viral infection to store viral proteins for degradation [49][50]. As studies progressed, these IBs more commonly became referred to as viral factories, viroplasms, and virosomes [51]. Because the term inclusion bodies is also widely used by biologists and biochemists to describe the insoluble aggregates formed by unfolded or misfolded proteins, it has been proposed to rename these structures or define them based on an identifiable marker [52]. While the field awaits consensus of a new nomenclature, the authors adopted the conventional terminology throughout the article.
2.2. The Formation
As a hallmark of infection by NNS RNA viruses, the formation of IBs is observed in the cytoplasm of cells infected by paramyxoviruses (e.g., measles virus (MeV) [53][54], mumps virus (MuV) [55], Nipah virus (NiV) [56], parainfluenza virus type 3 (PIV3) [57], and PIV5 [58]), pneumoviruses (e.g., RSV [59] and human metapneumovirus (HMPV) [60]), rhabdoviruses (e.g., RABV [61] and VSV [62]), and filoviruses (e.g., Marburg virus (MARV) [63] and EBOV [64]). These membrane-less supramolecular constructions are spherical when they are small during the early times after infection, and then progress to become heterogeneous in shape and size over time. In many cases, these cytoplasmic inclusion bodies serve as the site of viral replication by concentrating their RNA replication machinery [58][60][61][62][63][64][65].
Biomolecular condensates of proteins and nucleic acids can form a membrane-less organelle via LLPS. Whereas the exact criteria for defining LLPS are still evolving, and additional characteristics have been described, a general consensus in the field is that a LLPS compartment should fulfill the following three criteria: maintaining a spherical shape; fusing with each other after contact; and exchanging molecules with its surrounding environment [66]. Interestingly, several earlier studies including those of EBOV [64] and PIV3 [57] observed the fusion events between small IBs. In addition, Borna disease virus (BDV), a member of the bornaviruses characterized by persistent infection and replication in the nuclei of infected cells, forms nuclear inclusions that actively exchange viral proteins with the surrounding milieu [67]. Based on these observations, together with the fact that viral IBs do not have an enclosing membrane but do contain a high level of viral proteins and RNA as well, it has been tempting to link the formation of IBs to LLPS. Studies on RABV [68], VSV [69], and MeV [70] established that two IBs can fuse and subsequently relax to form a larger spherical IB, displaying hallmarks of surface tension, and that IBs show reversible molecular exchange with the surrounding cytosol after photobleaching. Collectively, these findings established that IBs of these viruses are liquid in nature and phase separate from the cytosol via LLPS.
Whether the formation of IBs with liquid properties represents a universal feature of NNS RNA viruses remains to be determined. Based on the spherical morphology of RSV IBs [52] and the diffusion of N protein observed for the pseudo IBs formed in transfected cells [71], it is very likely that RSV IBs are membrane-less liquid organelles. Moreover, evidence was provided that these IBs are sites of RSV replication. On the other hand, NiV generates two types of IBs, but the viral RNA synthesis appears to occur outside of these structures. One type of IB adopts spherical shape located in the perinuclear region, and the other type has a lamellar or square shape underneath the plasma membrane [56]. These two types of IBs arise independently; covalently-modified viral matrix M protein recruits the nucleocapsid to the plasma membrane and drives the formation of the plasma membrane-associated IBs. Based on these observations, it was proposed that the membrane-associated IBs are sites of virion assembly, whereas the perinuclear IBs are aggresome-like structures [56]. While fusion was detected between pseudo-perinuclear IBs formed in transfected cells, no fusion events have been observed between plasma-membrane associated IBs in both transfected and infected cells. Thus, further studies are needed to determine whether LLPS plays a role in the formation of either IBs formed by NiV.
A transfection approach in the absence of viral infection was used to identify the minimal viral components involved in the formation of RABV, VSV, MeV, and RSV IBs. A common theme that emerged from these results is that co-expression of N and P proteins in transfected cells is either sufficient (for RABV [68], MeV [70], and RSV [71]) or required (for VSV [69]) to generate structures morphologically similar to viral IBs. Moreover, in the case of RABV, MeV, and RSV, co-expression of N and P alone forms IB-like structures that exhibit liquid properties such as fusion/relaxation and/or molecule exchange. These findings were subsequently corroborated and extended by an in vitro reconstitution using MeV [72] and RSV [71] N and P recombinant proteins purified from E. coli. The transfection-based and in vitro systems further allow the mapping of the domains within N and P necessary for the IB formation. Not surprisingly, the domain of P mediating its interaction with N (PCTD for RABV, VSV, and RSV; XD for MeV) is required for the formation of IBs. In addition, the dimerization domain (for RABV and VSV) or tetramerization domain (for MeV and RSV) of P protein, and an intrinsic disorder region (IDR) of P (IDD2 for RABV, Ploop for MeV, and a.a. (160–227) for RSV), also play an important role. The oligomerization domain and IDR are two common protein structures contributing to the multivalent interactions underlying LLPS [19]. Taken together, these studies demonstrate that the nucleoproteins and phosphoproteins can be viewed as the basic scaffold for formation of the liquid-like IBs during NNS RNA virus infection. Interestingly, whereas expression of the nucleoprotein NP alone seems sufficient for EBOV IB formation with its C-terminal domain being necessary for this process, co-expression with the phosphoprotein VP35 can rescue the deletion of the C-terminal domain of NP to trigger IB formation [73]. Thus, even whether EBOV IBs are liquid organelles remains unknown, VP35 likely plays a role in the formation of IBs in vivo.
This entry is adapted from the peer-reviewed paper 10.3390/v13010126
References
- Koonin, E.V.; Dolja, V.V.; Krupovic, M.; Varsani, A.; Wolf, Y.I.; Yutin, N.; Zerbini, F.M.; Kuhn, J.H.; Global organization and proposed megataxonomy of the virus world. Microbiol. Mol. Biol. Rev. 2020, 84, 84, 10.1128/MMBR.00061-19.
- Bloom, S.; The foundations of virology: Discoverers and discoveries, inventors and inventions, developers and technologies. Emerg. Infect. Dis. 2013, 19, 693, 10.3201/eid1904.130054.
- Green, T.J.; Zhang, X.; Wertz, G.W.; Luo, M.; Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 2006, 313, 357-360, 10.1126/science.1126953.
- Albertini, A.A.V.; Wernimont, A.K.; Muziol, T.; Ravelli, R.B.G.; Clapier, C.R.; Schoehn, G.; Weissenhorn, W.; Ruigrok, R.W.H.; Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 2006, 313, 360-363, 10.1126/science.1125280.
- Tawar, R.G.; Duquerroy, S.; Vonrhein, C.; Varela, P.F.; Damier-Piolle, L.; Castagné, N.; MacLellan, K.; Bedouelle, H.; Bricogne, G.; Bhella, D.; et al. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 2009, 326, 1279-1283, 10.1126/science.1177634.
- Alayyoubi, M.; Leser, G.P.; Kors, C.A.; Lamb, R.A.; Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein–RNA complex. Proc. Natl. Acad. Sci. USA 2015, 112, E1792-E1799, 10.1073/pnas.1503941112.
- Sugita, Y.; Matsunami, H.; Kawaoka, Y.; Noda, T.; Wolf, M.; Cryo-EM structure of the Ebola virus nucleoprotein–RNA complex at 3.6 Å resolution. Nature 2018, 563, 137-140, 10.1038/s41586-018-0630-0.
- Shu, Y.; Habchi, J.; Costanzo, S.; Padilla, A.; Brunel, J.; Gerlier, D.; Oglesbee, M.; Longhi, S.; Plasticity in structural and functional interactions between the phosphoprotein and nucleoprotein of measles virus. J. Biol. Chem. 2012, 287, 11951-11967, 10.1074/jbc.m111.333088.
- Ding, H.; Green, T.J.; Lu, S.; Luo, M.; Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. J. Virol. 2006, 80, 2808-2814, 10.1128/jvi.80.6.2808-2814.2006.
- Brunel, J.; Chopy, D.; Dosnon, M.; Bloyet, L.-M.; Devaux, P.; Urzua, E.; Cattaneo, R.; Longhi, S.; Gerlier, D.; Sequence of events in measles virus replication: Role of phosphoprotein-nucleocapsid interactions. J. Virol. 2014, 88, 10851-10863, 10.1128/jvi.00664-14.
- Bruhn, J.F.; Barnett, K.C.; Bibby, J.; Thomas, J.M.H.; Keegan, R.M.; Rigden, D.J.; Bornholdt, Z.A.; Saphire, E.O.; Crystal structure of the Nipah virus phosphoprotein tetramerization domain. J. Virol. 2013, 88, 758-762, 10.1128/jvi.02294-13.
- Dias, A.; Bouvier, D.; Crépin, T.; McCarthy, A.A.; Hart, D.J.; Baudin, F.; Cusack, S.; Ruigrok, R.W.H.; The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 2009, 458, 914-918, 10.1038/nature07745.
- Reguera, J.; Weber, F.; Cusack, S.; Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog. 2010, 6, e1001101, 10.1371/journal.ppat.1001101.
- Morin, B.; Coutard, B.; Lelke, M.; Ferron, F.; Kerber, R.; Jamal, S.; Frangeul, A.; Baronti, C.; Charrel, R.; de Lamballerie, X.; et al. The N-terminal domain of the arenavirus L protein is an RNA endonuclease essential in mRNA transcription. PLoS Pathog. 2010, 6, e1001038, 10.1371/journal.ppat.1001038.
- Yuan, P.; Bartlam, M.; Lou, Z.; Chen, S.; Zhou, J.; He, X.; Lv, Z.; Ge, R.; Li, X.; Deng, T.; et al. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 2009, 458, 909-913, 10.1038/nature07720.
- Shin, Y.; Brangwynne, C.P.; Liquid phase condensation in cell physiology and disease. Science 2017, 357, eaaf4382, 10.1126/science.aaf4382.
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K.; Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285-298, 10.1038/nrm.2017.7.
- Boeynaems, S.; Alberti, S.; Fawzi, N.L.; Mittag, T.; Polymenidou, M.; Rousseau, F.; Schymkowitz, J.; Shorter, J.; Wolozin, B.; Van Den Bosch, L.; et al. Protein phase separation: A new phase in cell biology. Trends Cell Biol. 2018, 28, 420-435, 10.1016/j.tcb.2018.02.004.
- Gomes, E.; Shorter, J.; The molecular language of membraneless organelles. J. Biol. Chem. 2019, 294, 7115-7127, 10.1074/jbc.tm118.001192.
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Jülicher, F.; Hyman, A.A.; Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324, 1729-1732, 10.1126/science.1172046.
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J.; Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236-240, 10.1038/nature22822.
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H.; Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241-245, 10.1038/nature22989.
- Gibson, B.A.; Doolittle, L.K.; Schneider, M.W.G.; Jensen, L.E.; Gamarra, N.; Henry, L.; Gerlich, D.W.; Redding, S.; Rosen, M.K.; Organization of chromatin by intrinsic and regulated phase separation. Cell 2019, 179, 470-484.e21, 10.1016/j.cell.2019.08.037.
- Brangwynne, C.P.; Mitchison, T.J.; Hyman, A.A.; Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA 2011, 108, 4334-4339, 10.1073/pnas.1017150108.
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P.; Coexisting liquid phases underlie nucleolar subcompartments. Cell 2016, 165, 1686-1697, 10.1016/j.cell.2016.04.047.
- Handwerger, K.E.; Cordero, J.A.; Gall, J.G.; Cajal bodies, nucleoli, and speckles in the Xenopus oocyte nucleus have a low-density, sponge-like structure. Mol. Biol. Cell 2004, 16, 202-211, 10.1091/mbc.e04-08-0742.
- Corpet, A.; Kleijwegt, C.; Roubille, S.; Juillard, F.; Jacquet, K.; Texier, P.; Lomonte, P.; PML nuclear bodies and chromatin dynamics: Catch me if you can!. Nucleic Acids Res. 2020, 48, 11890-11912, 10.1093/nar/gkaa828.
- Marzahn, M.R.; Marada, S.; Lee, J.; Nourse, A.; Kenrick, S.; Zhao, H.; Ben-Nissan, G.; Kolaitis, R.-M.; Peters, J.L.; Pounds, S.; et al. Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J. 2016, 35, 1254-1275, 10.15252/embj.201593169.
- Kroschwald, S.; Maharana, S.; Mateju, D.; Malinovska, L.; Nüske, E.; Poser, I.; Richter, D.; Alberti, S.; Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife 2015, 4, e06807, 10.7554/elife.06807.
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P.; Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 2015, 163, 123-133, 10.1016/j.cell.2015.09.015.
- Patel, A.; Lee, H.O.; Jawerth, L.; Maharana, S.; Jahnel, M.; Hein, M.Y.; Stoynov, S.; Mahamid, J.; Saha, S.; Franzmann, T.M.; et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 2015, 162, 1066-1077, 10.1016/j.cell.2015.07.047.
- Jiang, H.; Wang, S.; Huang, Y.; He, X.; Cui, H.; Zhu, X.; Zheng, Y.; Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell 2015, 163, 108-122, 10.1016/j.cell.2015.08.010.
- Michnick, S.; Bergeron-Sandoval, L.-P.; Pappu, R.; François, P.; Hendricks, A.G.; Ehrlicher, A.J.; Heris, H.K.; A protein condensate drives actin-independent endocytosis. Biophys. J. 2019, 116, 161a, 10.1016/j.bpj.2018.11.894.
- Ouyang, M.; Li, X.; Zhang, J.; Feng, P.; Pu, H.; Kong, L.; Bai, Z.; Rong, L.; Xu, X.; Chi, W.; et al. Liquid-liquid phase transition drives intra-chloroplast cargo sorting. Cell 2020, 180, 1144-1159.e20, 10.1016/j.cell.2020.02.045.
- Fujioka, Y.; Alam, J.M.; Noshiro, D.; Mouri, K.; Ando, T.; Okada, Y.; May, A.I.; Knorr, R.L.; Suzuki, K.; Ohsumi, Y.; et al. Phase separation organizes the site of autophagosome formation. Nature 2020, 578, 301-305, 10.1038/s41586-020-1977-6.
- Boija, A.; Klein, I.A.; Sabari, B.R.; Dall’Agnese, A.; Coffey, E.L.; Zamudio, A.V.; Li, C.H.; Shrinivas, K.; Manteiga, J.C.; Hannett, N.M.; et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 2018, 175, 1842-1855.e16, 10.1016/j.cell.2018.10.042.
- Cho, W.-K.; Spille, J.-H.; Hecht, M.; Lee, C.; Li, C.; Grube, V.; Cisse, I.I.; Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018, 361, 412-415, 10.1126/science.aar4199.
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361, eaar3958, 10.1126/science.aar3958.
- Yasuda, S.; Tsuchiya, H.; Kaiho, A.; Guo, Q.; Ikeuchi, K.; Endo, A.; Arai, N.; Ohtake, F.; Murata, S.; Inada, T.; et al. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 2020, 578, 296-300, 10.1038/s41586-020-1982-9.
- Alberti, S.; Dormann, D.; Liquid–liquid phase separation in disease. Annu. Rev. Genet. 2019, 53, 171-194, 10.1146/annurev-genet-112618-043527.
- Boke, E.; Ruer, M.; Wühr, M.; Coughlin, M.; Lemaitre, R.; Gygi, S.P.; Alberti, S.; Drechsel, D.; Mitchison, T.J.; Amyloid-like self-assembly of a cellular compartment. Cell 2016, 166, 637-650, 10.1016/j.cell.2016.06.051.
- Frey, S.; Richter, R.P.; Görlich, D.; FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 2006, 314, 815-817, 10.1126/science.1132516.
- Schmidt, H.B.; Görlich, D.; Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. eLife 2015, 4, e04251, 10.7554/elife.04251.
- Alenquer, M.; Vale-Costa, S.; Etibor, T.A.; Ferreira, F.; Sousa, A.L.; Amorim, M.J.; Influenza A virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites. Nat. Commun. 2019, 10, 1629, 10.1038/s41467-019-09549-4.
- Amorim, M.J.; A comprehensive review on the interaction between the host GTPase Rab11 and influenza A virus. Front. Cell Dev. Biol. 2019, 6, 176, 10.3389/fcell.2018.00176.
- Pinkerton, H.; The morphology of viral inclusions and their practical importance in the diagnosis of human disease. Am. J. Clin. Pathol. 1950, 20, 201-207, 10.1093/ajcp/20.3.201.
- Albrecht, T.; Fons, M.; Boldogh, I.; Rabson, A.S. Effects on cells. In Medical Microbiology; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; ISBN 978-0-9631172-1-2.
- Fields, B.N.; Knipe, D.M.; Howley, P.M. Fields Virology; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; ISBN 978-1-4511-0563-6.
- Wileman, T.; Aggresomes and pericentriolar sites of virus assembly: Cellular defense or viral design?. Annu. Rev. Microbiol. 2007, 61, 149-167, 10.1146/annurev.micro.57.030502.090836.
- Novoa, R.R.; Calderita, G.; Arranz, R.; Fontana, J.; Granzow, H.; Risco, C.; Virus factories: Associations of cell organelles for viral replication and morphogenesis. Biol. Cell 2005, 97, 147-172, 10.1042/bc20040058.
- Netherton, C.; Moffat, K.; Brooks, E.; Wileman, T.; A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv. Virus Res. 2007, 70, 101-182, 10.1016/s0065-3527(07)70004-0.
- Jobe, F.; Simpson, J.; Hawes, P.; Guzman, E.; Bailey, D.; Respiratory syncytial virus sequesters NF-κb subunit p65 to cytoplasmic inclusion bodies to inhibit innate immune signaling. J. Virol. 2020, 94, e01380-20, 10.1128/jvi.01380-20.
- Pfaller, C.K.; Radeke, M.J.; Cattaneo, R.; Samuel, C.E.; Measles virus C protein impairs production of defective copyback double-stranded viral RNA and activation of protein kinase R. J. Virol. 2014, 88, 456-468, 10.1128/jvi.02572-13.
- Okonski, K.M.; Samuel, C.E.; Stress granule formation induced by measles virus is protein kinase PKR dependent and impaired by RNA adenosine deaminase ADAR1. J. Virol. 2013, 87, 756-766, 10.1128/jvi.02270-12.
- Katoh, H.; Kubota, T.; Kita, S.; Nakatsu, Y.; Aoki, N.; Mori, Y.; Maenaka, K.; Takeda, M.; Kidokoro, M.; Heat shock protein 70 regulates degradation of the mumps virus phosphoprotein via the ubiquitin-proteasome pathway. J. Virol. 2015, 89, 3188-3199, 10.1128/jvi.03343-14.
- Ringel, M.; Heiner, A.; Behner, L.; Halwe, S.; Sauerhering, L.; Becker, N.; Dietzel, E.; Sawatsky, B.; Maisner, A.; Nipah virus induces two inclusion body populations: Identification of novel inclusions at the plasma membrane. PLoS Pathog. 2019, 15, e1007733, 10.1371/journal.ppat.1007733.
- Zhang, S.; Jiang, Y.; Cheng, Q.; Zhong, Y.; Qin, Y.; Chen, M.; Inclusion body fusion of human parainfluenza virus type 3 regulated by acetylated α-tubulin enhances viral replication. J. Virol. 2017, 91, e01802-16, 10.1128/jvi.01802-16.
- Carlos, T.S.; Young, D.F.; Schneider, M.; Simas, J.P.; Randall, R.E.; Parainfluenza virus 5 genomes are located in viral cytoplasmic bodies whilst the virus dismantles the interferon-induced antiviral state of cells. J. Gen. Virol. 2009, 90, 2147-2156, 10.1099/vir.0.012047-0.
- Garcı́a, J.; Garcı́a-Barreno, B.; Vivo, A.; Melero, J.A.; Cytoplasmic inclusions of respiratory syncytial virus-infected cells: Formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology 1993, 195, 243-247, 10.1006/viro.1993.1366.
- Cifuentes-Muñoz, N.; Branttie, J.; Slaughter, K.B.; Dutch, R.E.; Human metapneumovirus induces formation of inclusion bodies for efficient genome replication and transcription. J. Virol. 2017, 91, e01282-17, 10.1128/jvi.01282-17.
- Lahaye, X.; Vidy, A.; Pomier, C.; Obiang, L.; Harper, F.; Gaudin, Y.; Blondel, D.; Functional characterization of negri bodies (NBs) in rabies virus-infected cells: Evidence that NBs are sites of viral transcription and replication. J. Virol. 2009, 83, 7948-7958, 10.1128/jvi.00554-09.
- Heinrich, B.S.; Cureton, D.K.; Rahmeh, A.A.; Whelan, S.P.J.; Protein expression redirects vesicular stomatitis virus RNA synthesis to cytoplasmic inclusions. PLoS Pathog. 2010, 6, e1000958, 10.1371/journal.ppat.1000958.
- Dolnik, O.; Stevermann, L.; Kolesnikova, L.; Becker, S.; Marburg virus inclusions: A virus-induced microcompartment and interface to multivesicular bodies and the late endosomal compartment. Eur. J. Cell Biol. 2015, 94, 323-331, 10.1016/j.ejcb.2015.05.006.
- Hoenen, T.; Shabman, R.S.; Groseth, A.; Herwig, A.; Weber, M.; Schudt, G.; Dolnik, O.; Basler, C.F.; Feldmann, H.; Inclusion bodies are a site of ebolavirus replication. J. Virol. 2012, 86, 11779-11788, 10.1128/jvi.01525-12.
- Rincheval, V.; Lelek, M.; Gault, E.; Bouillier, C.; Sitterlin, D.; Blouquit-Laye, S.; Galloux, M.; Zimmer, C.; Rameix-Welti, M.-A.; Functional organization of cytoplasmic inclusion bodies in cells infected by respiratory syncytial virus. Nat. Commun. 2017, 8, 563, 10.1038/s41467-017-00655-9.
- Alberti, S.; Gladfelter, A.; Mittag, T.; Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 2019, 176, 419-434, 10.1016/j.cell.2018.12.035.
- Charlier, C.M.; Wu, Y.-J.; Allart, S.; Malnou, C.E.; Schwemmle, M.; Gonzalez-Dunia, D.; Analysis of borna disease virus trafficking in live infected cells by using a virus encoding a tetracysteine-tagged P protein. J. Virol. 2013, 87, 12339-12348, 10.1128/jvi.01127-13.
- Nikolic, J.; Le Bars, R.; Lama, Z.; Scrima, N.; Lagaudrière-Gesbert, C.; Gaudin, Y.; Blondel, D.; Negri bodies are viral factories with properties of liquid organelles. Nat. Commun. 2017, 8, 58, 10.1038/s41467-017-00102-9.
- Heinrich, B.S.; Maliga, Z.; Stein, D.A.; Hyman, A.A.; Whelan, S.P.J.; Phase transitions drive the formation of vesicular stomatitis virus replication compartments. mBio 2018, 9, e02290-17, 10.1128/mbio.02290-17.
- Zhou, Y.; Su, J.M.; Samuel, C.E.; Ma, D.; Measles virus forms inclusion bodies with properties of liquid organelles. J. Virol. 2019, 93, e00948-19, 10.1128/jvi.00948-19.
- Galloux, M.; Risso-Ballester, J.; Richard, C.-A.; Fix, J.; Rameix-Welti, M.-A.; Eléouët, J.-F.; Minimal elements required for the formation of respiratory syncytial virus cytoplasmic inclusion bodies in vivo and in vitro. mBio 2020, 11, e01202-20, 10.1128/mbio.01202-20.
- Guseva, S.; Milles, S.; Jensen, M.R.; Salvi, N.; Kleman, J.-P.; Maurin, D.; Ruigrok, R.W.H.; Blackledge, M.; Measles virus nucleo- and phosphoproteins form liquid-like phase-separated compartments that promote nucleocapsid assembly. Sci. Adv. 2020, 6, eaaz7095, 10.1126/sciadv.aaz7095.
- Miyake, T.; Farley, C.M.; Neubauer, B.E.; Beddow, T.P.; Hoenen, T.; Engel, D.A.; Ebola virus inclusion body formation and RNA synthesis are controlled by a novel domain of nucleoprotein interacting with VP35. J. Virol. 2020, 94, e02100-19, 10.1128/jvi.02100-19.