Exosomes are endosome-derived nanovesicles produced by healthy as well as diseased cells. Their proteic, lipidic and nucleic acid composition is related to the cell of origin, and by vehiculating bioactive molecules they are involved in cell-to-cell signaling, both in healthy and pathologic conditions. Being nano-sized, non-toxic, biocompatible, scarcely immunogenic, and possessing targeting ability and organotropism, exosomes have been proposed as nanocarriers for their potential application in diagnosis and therapy. Among the different techniques exploited for exosome isolation, the sequential ultracentrifugation/ultrafiltration method seems to be the gold standard; alternatively, commercially available kits for exosome selective precipitation from cell culture media are frequently employed. To load a drug or a detectable agent into exosomes, endogenous or exogenous loading approaches have been developed, while surface engineering procedures, such as click chemistry, hydrophobic insertion and exosome display technology, allow for obtaining actively targeted exosomes.
- exosomes
- extracellular vesicles
- diagnosis
- theranostic
Please note: Below is an entry draft based on your previous paper, which is wrirren tightly around the entry title. Since it may not be very comprehensive, we kindly invite you to modify it (both title and content can be replaced) according to your extensive expertise. We believe this entry would be beneficial to generate more views for your work. In addition, no worry about the entry format, we will correct it and add references after the entry is online (you can also send a word file to us, and we will help you with submitting).
1. Introduction
All prokaryotic and eukaryotic cells secrete, in an evolutionary conserved way, extracellular vesicles (EVs), i.e., membrane-derived nano- and microvesicles [1]. For a long time, these vesicles were supposed to be either a waste removal system, products of cellular damage, or experimental artefacts [2]. Nowadays, EVs are recognized as specific cellular components performing different biological functions [3,4].
EVs are classified on the basis of the different sizes, cellular compartment of origin and localization, either inside or outside the cells [1]. Among them, exosomes, microparticles, shedding vesicles, apoptotic bodies, tolerosomes, prostasomes and prominosomes have been distinguished [5]. Two main processes for EV formation have been identified: some EVs, such as exosomes, apparently derive from exocytosis of multivesicular bodies, a part of the endosomal system including primary endocytic vesicles, early and late endosomes, and lysosomes [6]; otherwise, EVs may form from the direct budding of the cell membrane [2].
Exosomes, firstly identified by Johnestone in 1987 [7], represent a homogenous class of EVs in terms of size (30–150 nm), density (1.13–1.19 g/mL [8,9]) and membrane composition, differently from the other classes of EVs, which are characterized by higher heterogeneity [10,11]. As exosomes derive from the membrane of late endosomes [1], their proteome is particularly rich in tetraspanins (CD9, CD63, CD81 and CD82) and heat shock proteins (HSP70, HSP90), but also includes transmembrane proteins that are specific to the parent cell; for example, exosomes deriving from platelets contain P-selectin and intercellular adhesion molecule-1, while α- and β-chains of integrins are expressed on the membrane of exosomes deriving from dendritic cells, reticulocytes and T-cells [12]. As for exosome lipid composition, phosphatidylserine, sphingomyelin, cholesterol, ceramides, and ganglioside GM3 are particularly abundant [1]. Sharing the same biogenesis, exosomes and apoptotic bodies present analogous membrane topology, with the interior side of the vesicle corresponding to the cytosolic side of the parent cell membrane, although phosphatidylserine is specifically exposed on the outer leaflet of the vesicle membrane, due to the activity of enzymes such as flippase, floppase and scramblase [11]. Moreover, exosomes carry nucleic acids such as mRNA, siRNA, miRNA and DNA fragments.
2. Biological Function
By vehiculating proteins and genetic material, exosomes are involved in cell-to-cell communication by molecule transfer from donor to nearby, as well as distant, recipient cells [13,14]. Some authors have already reviewed the physiologic functions of exosomes in the immune system [15,16]; for instance, an in vitro study on both human and murine models evidenced that exosomes deriving from lymphocytes stimulate CD4+ T cell clones, revealing that these EVs might be involved in the transfer of peptidic signals among immune cells [8].
The exosomal mechanism of interaction with recipient cells has not been clarified yet (Figure 1). Some studies reported that exosomes fuse with the plasma membrane of the recipient cell, releasing their content in the cytoplasm [17,18]; as observed by Parolini et al. [19], the fusion process might be facilitated under acidic pH, typical of the tumor intracellular environment.
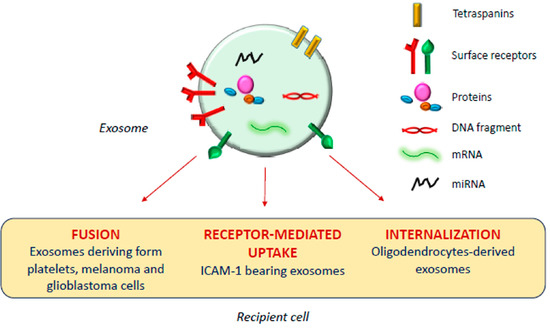
Actually, besides shuttling bioactive molecules among healthy cells, exosomes also play a crucial role under pathologic conditions such as cancer, and neurodegenerative, cardiovascular, infectious and respiratory diseases [14,20,21,22,23].
Tumor cells release high amounts of EVs, and several studies report a possible involvement of tumor-derived exosomes in different phases of tumor formation and progression, as reviewed in [24], although opposing pieces of evidence have been reported by different research groups. Al-Nedawi et al. [25] observed that glioblastoma cell-derived EVs membranes are enriched in a mutant form of the epidermal growth factor receptor, which promotes anti-apoptotic pathways and increases the anchorage-independent growth capacity; on the other hand, Ristorcelli et al. [26] evidenced that EVs secreted by pancreatic tumor cells, due to their high membrane content of cholesterol and sphingomyelin, induce activation of the mitochondrial apoptotic pathway in tumor cells, and Zitvogel and his group [27] even demonstrated the capability of dendritic cell-deriving exosomes to inhibit tumor growth in vivo.
Besides membrane composition, the exosomal content may also influence tumor progression. Some studies evidenced the role of miRNA present in tumor-derived exosomes in promoting neovascularization and angiogenesis [28,29]; also, a recent work of Gerloff et al. [30] evidenced that cutaneous melanoma-derived exosomes are enriched in miR-125b-5p, which induces a tumor-promoting phenotype in tumor-associated macrophages, while Kurahashi et al. [31] observed increased miR-204-5p levels in urinary exosomes of transgenic mice used as a model of a rare form of renal cancer.
An important aspect of tumor progression is the capacity of cells to elude the immune system, and several studies demonstrated that tumor-derived exosomes are involved in this process. Chalmin et al. [32] isolated exosomes deriving from different murine and human cancer cell lines and identified the interaction between Hsp-72, associated with the exosome membranes, and Stat3, expressed by the parent cells, as the key factor inducing the immunosuppressing activity of both mouse and human myeloid-derived suppressor cells.
Moreover, several papers highlighted the involvement of tumor-derived exosomes in the metastastic process [33,34,35,36]. For example, Peinado et al. [37] demonstrated that the highly metastatic behavior of primary melanomas might be ascribed to the abundant generation of exosomes influencing bone marrow progenitors, although different pieces of evidence were obtained in a replication study in 2018 [38]. Ramteke et al. [39] demonstrated that prostate cancer cell-derived exosomes, secreted under the hypoxic conditions typical of the malignancy, enhance the invasiveness of the tumor through induction of the cleavage of E-cadherin, a protein involved in the adherens junctions among epithelial cells. Another study [40] demonstrated that exosomes deriving from bladder cancer cells promote lymphatic metastasis through the action of a long non-coding RNA vehiculated by the vesicles; similarly, miR-105, miR-122 and miR-200-containing EVs promote breast cancer cell metastasis [41,42,43] and miR-221-containing exosomes derived from gastric cancer mesenchymal stem cells were found to promote migration of human gastric cancer cells in vitro [44].
3. Applications in Therapy
Exosomes are endowed with several characteristics suitable for drug delivery: they are nano-sized, non-toxic, biocompatible, scarcely immunogenic, and possess targeting ability and organotropism [45]. Indeed, exosomes are similar to small unilamellar liposomes in terms of size and capacity to carry both hydrophilic and lipophilic molecules, but the asymmetrical lipid distribution and specific protein composition of exosome membranes justify their organotropism and homing ability [46], confirmed by the evidence that cancer-derived exosomes fuse preferentially with their parent cells [47].
However, the clinical translation of exosomes as drug carriers is affected by several technical issues, including low production yield, considerable structural heterogeneity and complexity, difficulties in drug loading and in developing standard, scalable, and cost-effective GMP procedures for exosome isolation and purification [48]. To overcome these issues, bioinspired exosome-like vesicles have emerged as an alternative to naturally derived exosomes. Most of the artificial exosome-mimetic systems proposed to date stem from liposomes—the so-called hybrid exosomes derive from the fusion of exosome and liposome membranes [49]—or are obtained by serial extrusion of a parent cell suspension through decreasing pore size membranes [50].
Many research groups have developed exosomes and exosome-mimetic systems as nanocarriers for cancer treatment [45], proposing different techniques for vesicle isolation and purification, drug loading and surface functionalization [51,52]. The drug loading methods, in particular, can be classified in two main classes, endogenous and exogenous loading [52]. Endogenous loading includes the genetic modification of the parent cells, to have them to express specific proteins or nucleic acids to be included in the released vesicles, or their simple incubation with the drug to be loaded; exogenous loading implicates the incorporation of the drug in exosomes previously isolated from cell culture media or body fluids (urine, blood, saliva, breast milk, etc.).
Various active principles have been loaded into vesicles developed for the treatment of different types of cancer, such as doxorubicin [53,54], paclitaxel [55], gemcitabine [56], but also aspirin [57], imperialine [58], several miRNA [59,60,61,62,63,64] and mRNA molecules [65], tumor necrosis factor-α [66] and recombinant methioninase [67].
4. Applications in Diagnosis
Beyond their possible use as therapeutic active carriers, exosomes can be employed in the diagnostic field with two different approaches. Passive diagnostic applications involve the use of naturally derived tumor exosomes as cancer diagnostic and prognostic biomarkers [68,69], since, differently from circulating cancer cells, their abundance in blood allows for easy detection in small volumes of frozen plasma or serum [70]. By analyzing the proteomic and genomic profile of these exosomes, including mRNA, miRNA and mitochondrial RNA, it is possible to determine the type of tumor and its stage [68,71,72,73,74]. As an example, Zong et al. [75] developed silicon quantum dots (Si-QD) decorated with a CD63 aptamer to bind CD63 expressed on exosomes isolated from human breast SKBR3 cancer cells [12], thus obtaining a nanoprobe for super-resolution microscopy suitable for trafficking studies in live cells and for the investigation of exosome role in cancer metastasis. Moreover, Chen et al. [76] developed an exosome-based system for super-resolution microscopy, demonstrating the possibility of simultaneous dual-color imaging by immunofluorescent labeling of CD63 and HER2 molecules expressed on SKBR3-derived exosomes.
On the other hand, some researchers proposed to exploit exosomes as an active diagnostic tool, by manipulating them with compounds or nanoparticles (NP) detectable using different imaging techniques, such as optical fluorescence, computed tomography (CT), positron emission tomography (PET), single photon emission computed tomography (SPECT), and magnetic resonance imaging (MRI), for their use in the diagnosis of some forms of cancer that are difficult to reach, such as brain tumors, or in the early detection of cancer recurrences and metastasis, within a precision medicine approach [77].
On the wave of growing interest in exosome and EVs applications, the aim of this review is to summarize the diagnostic or theranostic platforms based on exosomes (Table 1) or exosome-mimetic vesicles (Table 2) that have been developed so far, classified on the basis of the labeling probes; in particular, the review focuses on the diverse manufacturing (Figure 2), loading and surface modification procedures, and on applications in oncology; the final paragraph briefly reports on exosome applications in other pathologic conditions.
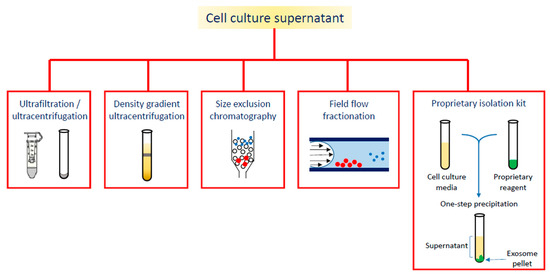
Figure 2. Different strategies for exosome isolation.
Table 1. Theranostic platforms based on exosomes, classified by loading strategy.
|
Ref. |
Labeling Strategy |
Parent Cells |
Exosome Isolation Method |
Labeling Compound |
Therapeutic Compound |
Loading/Labeling Procedure |
Surface Engineering |
Detection Technique |
Tests |
|
[78] |
Nanoparticle-loaded exosomes |
Raw264.7 mouse macrophages |
Sequential centrifugation |
SPION |
Curcumin |
Exogenous (electroporation) |
NRP-1 binding peptide by click chemistry |
MRI |
In vitro: U251 cells In vivo: BALB/c nude mice transplanted with U251 cells |
|
[79] |
SKBR3 breast cancer cells |
Exosome isolation kit |
Gold-carbon QD |
Exogenous (incubation exploiting targeted loading through anti-HER2 antibodies) |
Fluorescence imaging |
In vitro: HeLa cells |
|||
|
[80] |
MCF-7 breast cancer cells |
Exosome isolation kit |
Vanadium carbide QD |
Exogenous (electroporation) |
RGD peptide introduced by incubating exosomes with DSPE-PEG-RGD |
Photoacoustic imaging |
In vitro: MCF-7, A549, NHDF cells In vivo: tumor-bearing BALB/c nude mice |
||
|
[81] |
Urine of gastric cancer patients |
Sequential centrifugation |
Chlorine-6 labeled gold NP |
Exogenous (electroporation) |
Fluorescence imaging |
In vitro: MGC-803, Raw264.7 cells In vivo: MGC-803 tumor-bearing BALB/c-nude mice |
|||
|
[82,83] |
Murine adipose stem cells |
Exosome isolation kit |
USPION |
|
Endogenous (cell incubation) |
|
MRI |
In vitro: exosomes immobilized in an agarose matrix In vivo: C57BL/6 mice |
|
|
[84] |
Mesenchymal stem cells |
Sequential centrifugation |
Gold NP |
Exogenous (incubation) |
CT |
In vivo: C57bl/6 mice |
|||
|
[85] |
Transition metal-labeled exosomes |
Human umbilical cord mesenchymal stem cells |
Sequential centrifugation |
68Gd (complexed by DOTA) |
|
Exogenous (lipid insertion technique with Gd-DOTA-DSPE) |
|
MRI |
In vitro: K7M2 mouse and 14B human osteosarcoma cells In vivo: immunodeficient NU/NU nude mice implanted with K7M2 cells |
|
[86] |
Human umbilical cord blood mononuclear cells |
Sequential centrifugation |
64Cu (complexed by DOTA) |
|
Exogenous (reaction between the maleimide group of DOTA and thiol groups on exosome surface) |
|
PET/MRI |
In vitro: HUVEC In vivo: C57BL/6J mice |
|
|
[87] |
4T1 breast cancer cells |
Sequential centrifugation |
64Cu (complexed by NOTA) |
|
Exogenous (reaction of NOTA with exosome surface proteins) |
PEG decoration using PEG5k/NHS |
PET |
In vivo: 4T1 tumor-bearing BALB/c mice |
|
|
[88] |
Mouse macrophage HEK293T cells |
Sequential centrifugation |
99mTc |
Exogenous (incubation with fac-[99mTc(CO)3(H2O)3]+) |
DARPin G3 functionalization by transfection of the parent cells |
Radioactive signal by gamma-counter |
In vitro: SKOV-3, MCF-7, U87-MG, HT-29, A549 cells In vivo: BALB/c mice, SKOV-3 xenografted C57 nude mice |
||
|
[89] |
Human embryonic kidney HEK293 cells |
Sequential centrifugation |
111In |
Exogenous (incubation with 111In -oxine) |
CSPGAKVRC peptide, functionalized by transfection of the parent cells |
CT/SPECT |
In vitro: Raw264.7 cells In vivo: 4T1 tumor-bearing Balb/c mice |
||
|
[90] |
Bioluminescently labeled exosomes |
Human embryonic kidney 293T cells |
Sequential centrifugation |
Gaussia princeps luciferase (Gluc) |
Endogenous (transfection of the parent cells with a gene encoding for Gluc bound to a membrane protein) |
IVIS imaging |
In vivo: immunodeficient athymic nude mice |
||
|
[91] |
Human embryonic kidney 293T cells |
Sequential centrifugation |
GFP, tandem dimer Tomato |
Endogenous (transfection of the parent cells with a gene encoding for palmGFP/palmtdTomato) |
Multiphoton intravital microscopy |
In vitro: 293T cells In vivo: C57BL6 (B6) mice implanted with mouse thymoma EL-4 cells |
|||
|
[92] |
Nanocluster loaded exosomes |
HepG2 human hepatocellular carcinoma |
Sequential centrifugation |
Ag-nanoclusters and Fe3O4 NP |
Endogenous (parent cells cultured in the presence of AgNO3 and FeCl2 forming the nanoclusters) |
Flurescence bioimaging, CT, MRI |
In vitro: HepG2, U87 cells |
||
|
[93] |
Metabolic labeled exosomes |
MDA-MB-231 breast cancer cells |
Ultracentrifugation and size exclusion chromatography |
Deuterium |
Endogenous (parent cells cultured in presence of D2O/d-Gluc/d-Chol) |
Raman spectroscopic imaging |
In vitro: MDA-MB-231, MCF10A cells |
Table 2. Theranostic platforms based on exosome-mimetic vesicles.
|
Ref. |
Cell Line |
Labeling Compound |
Therapeutic Compound |
Vesicle Preparation Method |
Loading/Labeling Procedure |
Detection Technique |
Tests |
|
[94] |
Bel-7402 human hepatoma cancer cells |
NP-encapsulated doxorubicin |
NP-encapsulated doxorubicin |
Coating of the NP with cell membranes through extrusion |
Incubation |
Fluorescence imaging |
In vitro: Bel-7402, MCF-7, L-O2 cells |
|
[95] |
J774A.1 mouse macrophages |
Gd-conjugated liposomes |
|
Sonication and extrusion of the exosome/liposome mixture |
Obtained during vesicle preparation procedure |
MRI |
In vitro: K7M2, NIH/3T3 cells In vivo: osteosarcoma—bearing NU/NU immunodeficient mice |
|
[96] |
Raw264.7 mouse macrophages, HB1.F3 human neural stem cells |
99mTc-HMPAO |
Sequential extrusion of parent cells and density gradient centrifugation |
Incubation |
SPECT/CT |
In vivo: BALB/c mice |
This entry is adapted from the peer-reviewed paper 10.3390/cells9122569
