The packaging of the eukaryotic genome into chromatin regulates the storage of genetic information, including the access of the cell's DNA metabolism machinery. Indeed, since the processes of DNA replication, translation, and repair require access to the underlying DNA, several mechanisms, both active and passive, have evolved by which chromatin structure can be regulated and modified. One mechanism relies upon the function of chromatin remodeling enzymes which couple the free energy obtained from the binding and hydrolysis of ATP to the mechanical work of repositioning and rearranging nucleosomes. Here we review recent work on the nucleosome mobilization activity of this essential family of molecular machines.
- chromatin remodelers,nucleosome repositioning,molecular machines,enzyme mechanism
1. Introduction
The compaction of the eukaryotic genome into chromatin begins with the wrapping (i.e., bending) of the DNA around octamers of the H2A, H2B, H3, and H4 histone proteins to form nucleosomes[1][2][3][4]; two copies of each of these histones are found within each octamer. The DNA wrapped within each nucleosome, referred to as nucleosomal DNA, contacts the histone octamer at 14 different sites, spaced approximately 10 basepairs apart, each of which contains several different types of noncovalent interactions (e.g., van der Waals interactions, hydrogen bonds) between the DNA and the histones[1][3]. The histone octamer together with the ≈147 basepairs of nucleosomal DNA wrapped around it is referred to as the nucleosome core particle (NCP). A single base pair of the nucleosomal DNA is centered on the nucleosome dyad and defines the pseudo-two-fold symmetry axis of the NCP[5]. DNA locations are designated by superhelical locations (SHL) representing superhelical turns from the dyad (SHL0) and ranging from SHL−7 to SHL7[5][6]. As shown in Figure 1,the next level of chromatin packing is the ordering of nucleosomes into a “beads on a string” structure in which individual nucleosomes are separated by short stretches of free DNA, flanking either side of the NCP, that link NCPs together[7]; this free DNA is most often referred to as linker DNA, but is sometimes called flanking DNA.
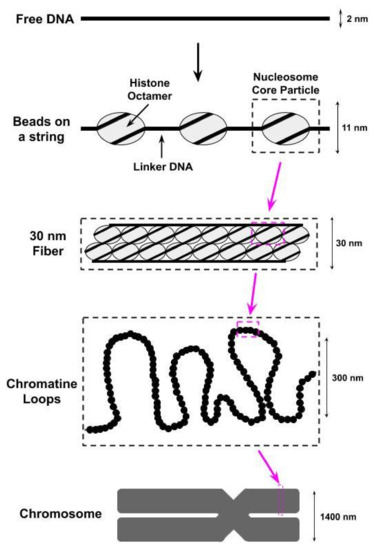
Figure 1. The first level of compacting the genome into chromatin involves wrapping the DNA around octamers of histone proteins to form nucleosomes. We identify in this figure a nucleosome core particle, which consists of an octamer of histone proteins wrapped by ≈147 basepairs of DNA. These nucleosome core particles are further wrapped together to form a structure known as a 30 nm fiber due to its physical dimensions. Supercoiling of the 30 nm fibers into chromatine loops then leads to the formation of a chromosome.
Since the packaging of DNA within nucleosomes requires that the DNA be wrapped around the histone octamer and that specific contacts be made between the DNA and the histones[1][2][3][4][8], it is not surprising that the thermodynamic stability of a nucleosome depends upon the sequence of the wrapped DNA[9][10][11][12][13]; however, DNA sequence has little affect on nucleosome structure[2]. For example, sequences of DNA that are more easily bent display a higher affinity for octamer binding[14] and nucleosomes reconstituted using such DNA sequences are also more stable[14]. The periodicity of particular dinucleotides in DNA sequences with high affinity for histone octamer binding further demonstrates the correlation between DNA sequence and nucleosome stability as the positions of these dinucleotides in the DNA increase the flexibility of the DNA, thereby facilitating the wrapping the DNA around the histone octamer[9][10][14]. To that point, it is worth noting that in several studies of nucleosome structure, nucleosomal DNA accessibility, and chromatin remodeling activity, the NCP substrates were reconstituted using a sequence of DNA, referred to as the Widom 601 sequence, that was designed to have high affinity for nucleosome binding[9].
The wrapping of nucleosomal DNA around the histone octamer restricts the ability of DNA binding proteins to access it[15][16][17]. Consequently, the wrapping of nucleosomal DNA must be dynamically controlled in order to regulate the accessibility of DNA repair, DNA replication, and gene expression machinery[18][19]. One mechanism of control involves the activity of molecular motors called chromatin remodelers, which reposition NCPs along DNA (i.e., move histone octamers relative to the DNA) using an ATP-dependent mechanism[20][21][22]. Chromatin remodelers have been shown to play a role in many biological processes, from regulation of gene expression to DNA damage response, and to catalyze many enzymatic reactions, including nucleosome assembly and disassembly [20][23][24][25][26][27].
2. Mechanisms of Nucleosome Repositioning
Nucleosome repositioning is an important part of the epigenetic regulation of gene expression. Chromatin remodelers modify nucleosome spacing through the sliding of NCPs relative to the DNA or by removing/exchanging histones from the NCP[28]. The former process relies upon the ability of the chromatin remodeler to translocate the nucleosomal DNA. Indeed, chromatin remodelers share the ATP-dependent DNA translocation activity of other members of the helicase superfamily SF2 [23][29][30][31][32][33][34]. Chromatin remodelers lack the helicase activity [35] of other SF2 enzymes, however, since their ATPase domain lacks the wedge domain necessary for DNA strand separation[36].
The isolated ATPase subunit of the ISWI chromatin remodeler family has been shown to be an ATP-dependent DNA translocase as well as a functional chromatin remodeler [30][34][37]. Single-stranded DNA translocation by the ISWI ATPase has a 3′ to 5′ directional bias and low processivity[30][34]. This low processivity may result from an allosteric regulation of DNA binding by nucleotide binding as the binding of ATP reduces the DNA binding affinity of ISWI[38]. In contrast, NCP binding by ISWI shows much higher affinity and is not allosterically regulated by nucleotide binding [38][37]. These results thus provide further evidence that the HAND, SANT, and SLIDE domains share additional histone recognition responsibilities beyond their reported role in DNA binding[39]. It is also interesting that, although ISWI is a poorly processive DNA translocase, it is nevertheless remarkably efficient, moving an average of 14 nucleotides per ATP molecule hydrolyzed[40]. It should be noted, however, that non-uniform motion may inflate the estimate of this distance[40]. Regardless, DNA translocation by ISWI is more efficient than the 1–4 nucleotides translocated per ATP molecule hydrolyzed that has been reported for genetically related helicases[41][42][43].
The coupling of ATP hydrolysis to the DNA translocation activity of a minimal construct of the essential SWI/SNF-family RSC chromatin remodeler, consisting of three of the fifteen subunits of the full length RSC[44][45], has also been characterized[46]. This minimal construct, denoted as RSCt, consists of the ARP7 and ARP9 subunits together with a truncated version of the Sth1 subunit, which contains the DNA-binding ATPase and translocation motor [45][47]. Similar to what has been observed with ISWI, RSCt displays a 3′ to 5′ directional bias for DNA translocation[48][49] and low translocation processivity[46]. In contrast to what was observed for ISWI, however, translocation by RSCt is not as energy efficient as DNA translocation by ISWI or genetically related helicases[41][42][43], requiring the hydrolysis of three ATP molecules for each nucleotide translocated[46]. RSCt was also shown to undergo an initiation process following its binding to DNA before becoming competent for DNA translocation and to be capable of exerting enough force during DNA translocation to disrupt a biotin–streptavidin linkage[46].
It is also interesting to note that the macroscopic rates of DNA translocation by ISWI and RSCt are similar to the macroscpoic rate of DNA translocation by the NS3h helicase from hepatitis C virus[42], but much slower than the macroscopic rates of DNA translocation by the UvrD[50], Rep[51], and T7[41] helicases. This may suggest an underlying difference between helicase superfamilies with SFII helicases (NS3h, RSC, and ISWI) translocating along DNA much more slowly than SFI (UvrD and Rep) or DNAB-like (T7) helicases. Of course, while these results are interesting from the standpoint of making general comparisons, it is difficult to draw distinctive conclusions from them as these studies of these enzymes were performed under different conditions.
DNA translocation by remodelers does play a central role in many models of their nucleosome repositioning activity [39][52]. For example, the ATPase subunit of the Saccharomyces cerevisiae SWI/SNF and RSC chromatin remodelers are proposed to bind to a specific site within the NCP and subsequently engage in directional DNA translocation, pulling in DNA from one side of the NCP and pumping it out from the other side, to shift the histone octamer relative to the DNA[53][54]; this DNA translocation proceeds through the formation and propagation of DNA loops [48][54][55], twist defects in the DNA[56][57], or some combination of both (Figure 2). Thus, the SWI/SNF complex initiates nucleosome repositioning by competing with the histone octamer for DNA binding. The proposal that DNA loops are an intermediate in the nucleosome repositioning mechanism of SWI/SNF is further supported by observations that Saccharomyces cerevisiae RSC forms DNA loops during its translocation along free DNA [46][58]and with the formation of DNA loops in Snf-nucleosome complexes being allosterically regulated through nucleotide binding by Snf2[59].
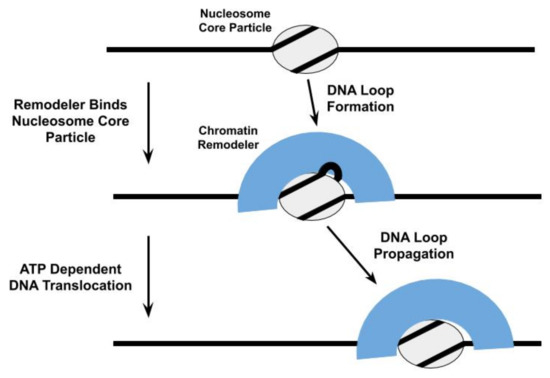
Figure 3. One model proposed for the repositioning of the nucleosome core particle involves the formation and propagation of DNA loops [60][[61][62][60].
DNA translocation plays a similar role in models of nucleosome repositioning by ISWI. Based upon single-molecule experiments, a “power-stroke” model has been proposed for the DNA translocation associated with nucleosome repositioning by Saccharomyces cerevisiae Isw1 and Isw2[52]. According to this model, the chromatin remodelers slide the DNA relative to the histone octamer through a coordinated mechanism controlling DNA entering or exiting the NCP[52]. The chromatin remodeler is bound to the linking DNA on either side of the NCP and initiates the repositioning reaction by translocating the nucleosomal DNA one basepair at a time toward one end of the NCP through an ATP-dependent mechanism, building up strain within the NCP between the DNA and the histones. After this strain becomes sufficiently strong, corresponding to seven basepairs of translocated nucleosomal DNA, a conformational change occurs between the linker-DNA-binding domain of the chromatin remodeler (possibly the SLIDE domain) and the ATPase domain, which pushes three basepairs of DNA into the NCP. This allows three basepairs of DNA to move outside the NCP from the other side of the NCP. This process then repeats in units of three basepairs translocated per translocation cycle. This model is consistent with the results of more recent ensemble experiments[63], theoretical analysis[64], and with the formation of DNA loops in Isw1-nucleosome complexes being allosterically regulated through nucleotide binding by Saccharomyces cerevisiae Isw1[60]. In an alternative ‘ratchet’ model for nucleosome repositioning by ISWI, the ATPase domain of the ISWI complex is responsible for all mechanical processes associated with nucleosome repositioning[37]. Remodeling still originates from the seven base pairs of flanking DNA being pulled through the nucleosome by the ATPase domain which causes internal tensions that must be resolved through the breaking and reforming of connections between the histones and the nucleosomal DNA[37]. This model assumes that the HAND, SANT, and SLIDE domains are responsible for passive histone recognition and the removal of the NegC domain from the ATPase cleft[37][65][66]. In each of these two models for nucleosome repositioning by ISWI, multiple rounds of ATP hydrolysis are required to reposition the nucleosome. Although additional work is required to further discriminate between these two models, it is worth noting that the step-size observed for DNA translocation by isolated ATPase subunit of Xenopus laevis ISWI is similar to what is predicted for the initial step in the ‘ratchet’ model[40]. Lastly, the Saccharomyces cerevisiae INO80 chromatin remodeler is capable of moving an NCP from an end position to a more central position along DNA and may serve as a nucleosome spacing factor[67]. Although it has been proposed that NCP binding by INO80 depends upon the length of the DNA flanking the NCP[67], that assessment was made without a concomitant determination of the stoichiometry of INO80 binding to the NCP. Indeed, it is possible that the changes in the apparent Kd of NCP binding by INO80 associated with increasing length of flanking DNA may be attributed to additional biding sites on the flanking DNA itself, as was observed for ISWI[38].
Although ATPase activity is required for remodeler function, several studies have shown that the movement of the histone octamer relative to the DNA is poorly coupled to ATP hydrolysis for Drosophila ACF[68], human Snf2H[69], and Xenopus laevis ISWI[63]. This poor coupling efficiency between ATPase activity and octamer movement could be a fundamental property of these motors (i.e., these motors are inherently inefficient at nucleosome repositioning) or it could result from significant ATP hydrolysis being associated with either futile repositioning or the initiation of repositioning. A large energetic cost for initiating the repositioning reaction seems unlikely as the bulges of DNA suggested to be the precursors of DNA translocation and nucleosome repositioning can be formed by chromatin remodelers binding to nucleosomes in the absence of ATP hydrolysis[62][60]. Since DNA translocation is fundamental to most models of nucleosome repositioning, DNA translocation has been proposed to be the energetically rate-limiting process during nucleosome repositioning[70]. However, since DNA translocation by chromatin remodelers is remarkably energy efficient[40], it seems unlikely that DNA translocation itself would be significantly less energy inefficient when coupled to nucleosome repositioning. However, since nucleosome repositioning requires the chromatin remodeler to break and reform contacts between the histones and the nucleosomal DNA in order for DNA translocation to occur, disrupting the interactions between the histones and the nucleosomal DNA does provide an energetic barrier to nucleosome repositioning. This barrier, if sufficiently large, could result in multiple rounds of unsuccessful attempts at nucleosome repositioning and the associated DNA translocation for each successful repositioning event[63][64]. This hypothesis could also explain the poor template commitment reported for human Snf2H [69] as well as the observation that Xenopus laevis ISWI repositions nucleosomes through a random walk mechanism[63]. Indeed, since most studies of chromatin remodeler function involve NCP substrates reconstituted using the Widom 601 DNA sequence[9], it is possible that experiments intended to assess the repositioning activity of chromatin remodelers, may instead report primarily on the affinity of histone:DNA interactions within the nucleosome rather than the activity of the remodeler. This proposition is further supported by the observation that the rate of nucleosome repositioning by Xenopus laevis ISWI depends upon the sequence of the nucleosomal DNA, with faster repositioning occurring with DNA sequences with lower affinity for histone binding[64]. Indeed, if the DNA translocation associated with nucleosome repositioning requires the chromatin remodeler to compete with the histones for binding the nucleosomal DNA [60][61][54], then it should be more difficult for chromatin remodelers to reposition nucleosomes reconstituted with DNA sequences with higher affinity for histone binding[64]. It is therefore not surprising that several studies have shown that the sequence of nucleosomal DNA influences the nucleosome repositioning activity of chromatin remodelers [63][71][72][73][74]. It is worth noting that the nucleosome repositioning activity of Saccharomyces cerevisiae INO80 has been reported to be independent of the sequence of nucleosomal DNA[75], but a lack of determination of the stoichiometry of INO80 binding to the NCP substrates used in these experiments creates some ambiguity in the interpretation of those results, especially for nucleosome substrates containing long lengths of DNA flanking the NCP[38][48]. While a dependence of nucleosome repositioning on the sequence of nucleosomal DNA may be helpful in providing an alternative tool for measuring DNA–histone interactions (i.e., an alternative probe of histone:DNA interaction free energy[10]), it clearly muddles what can be concluded about the intrinsic repositioning activity of the remodeler and how this activity is influenced and modulated [64].
This entry is adapted from the peer-reviewed paper 10.3390/ijms22010076
References
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature 1997, 389, 251.
- Davey, C.A.; Sargent, D.F.; Luger, K.; Maeder, A.W.; Richmond, T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002, 319, 1097–1113.
- Richmond, T.J.; Davey, C.A. The structure of DNA in the nucleosome core. Nature 2003, 423, 145.
- Frouws, T.D.; Duda, S.C.; Richmond, T.J. X-ray structure of the MMTV-A nucleosome core. Proc. Natl. Acad. Sci. USA 2016, 113, 1214–1219.
- Flaus, A.; Luger, K.; Tan, S.; Richmond, T.J. Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc. Natl. Acad. Sci. USA 1996, 93, 1370–1375.
- McGinty, R.K.; Tan, S. Nucleosome structure and function. Chem. Rev. 2015, 115, 2255–2273.
- Szerlong, H.J.; Hansen, J.C. Nucleosome distribution and linker DNA: connecting nuclear function to dynamic chromatin structure. Biochem. Cell Biol. 2011, 89, 24–34.
- Dann, G.P.; Liszczak, G.P.; Bagert, J.D.; Müller, M.M.; Nguyen, U.T.; Wojcik, F.; Brown, Z.Z.; Bos, J.; Panchenko, T.; Pihl, R.; et al. ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature 2017, 548, 607–611.
- Lowary, P.; Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998, 276, 19–42.
- Thåström, A.; Lowary, P.; Widom, J. Measurement of histone–DNA interaction free energy in nucleosomes. Methods 2004, 33, 33–44.
- Battistini, F.; Hunter, C.A.; Moore, I.K.; Widom, J. Structure-based identification of new high-affinity nucleosome binding sequences. J. Mol. Biol. 2012, 420, 8–16.
- Ngo, T.T.; Yoo, J.; Dai, Q.; Zhang, Q.; He, C.; Aksimentiev, A.; Ha, T. Effects of cytosine modifications on DNA flexibility and nucleosome mechanical stability. Nat. Commun. 2016, 7, 1–9.
- Tóth, K.; Böhm, V.; Sellmann, C.; Danner, M.; Hanne, J.; Berg, M.; Barz, I.; Gansen, A.; Langowski, J. Histone-and DNA sequence-dependent stability of nucleosomes studied by single-pair FRET. Cytom. Part A 2013, 83, 839–846.
- Widom, J. Role of DNA sequence in nucleosome stability and dynamics. Q. Rev. Biophys. 2001, 34, 269–324.
- Polach, K.; Widom, J. Mechanism of protein access to specific DNA sequences in chromatin: A dynamic equilibrium model for gene regulation. J. Mol. Biol. 1995, 254, 130–149.
- Anderson, J.; Widom, J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 2000, 296, 979–987.
- Li, G.; Widom, J. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 2004, 11, 763–769.
- Azmi, I.F.; Watanabe, S.; Maloney, M.F.; Kang, S.; Belsky, J.A.; MacAlpine, D.M.; Peterson, C.L.; Bell, S.P. Nucleosomes influence multiple steps during replication initiation. eLife 2017, 6, e22512.
- Kadonaga, J.T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell 1998, 92, 307–313.
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304.
- Bowman, G.D. Mechanisms of ATP-dependent nucleosome sliding. Curr. Opin. Struct. Biol. 2010, 20, 73–81.
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422.
- Eisen, J.A.; Sweder, K.S.; Hanawalt, P.C. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995, 23, 2715–2723.
- Lusser, A.; Kadonaga, J.T. Chromatin remodeling by ATP-dependent molecular machines. Bioessays 2003, 25, 1192–1200.
- Tyagi, M.; Imam, N.; Verma, K.; Patel, A.K. Chromatin remodelers: We are the drivers! Nucleus 2016, 7, 388–404.
- Markert, J.; Luger, K. Nucleosomes Meet Their Remodeler Match. Trends Biochem. Sci. 2020.
- Peng, A.Y.T.; Kolhe, J.A.; Behrens, L.D.; Freeman, B.C. Genome organization: Tag it, move it, place it. Current Opinion in Cell Biology 2020, 68, 90–97.
- Narlikar, G.J.; Sundaramoorthy, R.; Owen-Hughes, T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 2013, 154, 490–503.
- Saha, A.; Wittmeyer, J.; Cairns, B.R. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002, 16, 2120–2134.
- Whitehouse, I.; Stockdale, C.; Flaus, A.; Szczelkun, M.D.; Owen-Hughes, T. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol. Cell. Biol. 2003, 23, 1935–1945.
- Zhang, Y.; Smith, C.L.; Saha, A.; Grill, S.W.; Mihardja, S.; Smith, S.B.; Cairns, B.R.; Peterson, C.L.; Bustamante, C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol. Cell 2006, 24, 559–568.
- Lia, G.; Indrieri, M.; Owen-Hughes, T.; Finzi, L.; Podesta, A.; Milani, P.; Dunlap, D. ATP-dependent looping of DNA by ISWI. J. Biophotonics 2008, 1, 280–286.
- Eastlund, A.; Malik, S.S.; Fischer, C.J. Kinetic mechanism of DNA translocation by the RSC molecular motor. Arch. Biochem. Biophys. 2013, 532, 73–83.
- Eastlund, A.; Al-Ani, G.; Fischer, C.J. Low processivity for DNA translocation by the ISWI molecular motor. Biochim. Biophys. Acta-(Bba)-Proteins Proteom. 2015, 1854, 1487–1493.
- Côté, J.; Peterson, C.L.; Workman, J.L. Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc. Natl. Acad. Sci. USA 1998, 95, 4947–4952.
- Dürr, H.; Körner, C.; Müller, M.; Hickmann, V.; Hopfner, K.P. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell 2005, 121, 363–373.
- Mueller-Planitz, F.; Klinker, H.; Ludwigsen, J.; Becker, P.B. The ATPase domain of ISWI is an autonomous nucleosome remodeling machine. Nat. Struct. Mol. Biol. 2013, 20, 82–89.
- Al-Ani, G.; Briggs, K.; Malik, S.S.; Conner, M.; Azuma, Y.; Fischer, C.J. Quantitative determination of binding of ISWI to nucleosomes and DNA shows allosteric regulation of DNA binding by nucleotides. Biochemistry 2014, 53, 4334–4345.
- Mueller-Planitz, F.; Klinker, H.; Becker, P.B. Nucleosome sliding mechanisms: new twists in a looped history. Nat. Struct. Mol. Biol. 2013, 20, 1026–1032.
- Eastlund, A.; Al-Ani, G.; Fischer, C.J. Low processivity for DNA translocation by the ISWI molecular motor. Biochim. Biophys. Acta-(Bba)-Proteins Proteom. 2015, 1854, 1487–1493.
- Kim, D.E.; Narayan, M.; Patel, S.S. T7 DNA helicase: a molecular motor that processively and unidirectionally translocates along single-stranded DNA. J. Mol. Biol. 2002, 321, 807–819.
- Khaki, A.R.; Field, C.; Malik, S.; Niedziela-Majka, A.; Leavitt, S.A.; Wang, R.; Hung, M.; Sakowicz, R.; Brendza, K.M.; Fischer, C.J. The macroscopic rate of nucleic acid translocation by hepatitis C virus helicase NS3h is dependent on both sugar and base moieties. J. Mol. Biol. 2010, 400, 354–378.
- Tomko, E.J.; Fischer, C.J.; Lohman, T.M. Single-stranded DNA translocation of E. coli UvrD monomer is tightly coupled to ATP hydrolysis. J. Mol. Biol. 2012, 418, 32–46.
- Sirinakis, G.; Clapier, C.R.; Gao, Y.; Viswanathan, R.; Cairns, B.R.; Zhang, Y. The RSC chromatin remodelling ATPase translocates DNA with high force and small step size. EMBO J. 2011, 30, 2364–2372.
- Malik, S.S.; Rich, E.; Viswanathan, R.; Cairns, B.R.; Fischer, C.J. Allosteric interactions of DNA and nucleotides with S. cerevisiae RSC. Biochemistry 2011, 50, 7881–7890.
- Eastlund, A.; Malik, S.S.; Fischer, C.J. Kinetic mechanism of DNA translocation by the RSC molecular motor. Arch. Biochem. Biophys. 2013, 532, 73–83.
- Cairns, B.R.; Erdjument-Bromage, H.; Tempst, P.; Winston, F.; Kornberg, R.D. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol. Cell 1998, 2, 639–651.
- Saha, A.; Wittmeyer, J.; Cairns, B.R. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002, 16, 2120–2134.
- Zhang, Y.; Smith, C.L.; Saha, A.; Grill, S.W.; Mihardja, S.; Smith, S.B.; Cairns, B.R.; Peterson, C.L.; Bustamante, C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol. Cell 2006, 24, 559–568.
- Fischer, C.J.; Maluf, N.K.; Lohman, T.M. Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J. Mol. Biol. 2004, 344, 1287–1309.
- Brendza, K.M.; Cheng, W.; Fischer, C.J.; Chesnik, M.A.; Niedziela-Majka, A.; Lohman, T.M. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc. Natl. Acad. Sci. USA 2005, 102, 10076–10081.
- Deindl, S.; Hwang, W.L.; Hota, S.K.; Blosser, T.R.; Prasad, P.; Bartholomew, B.; Zhuang, X. ISWI remodelers slide nucleosomes with coordinated multi-base-pair entry steps and single-base-pair exit steps. Cell 2013, 152, 442–452.
- Dechassa, M.L.; Zhang, B.; Horowitz-Scherer, R.; Persinger, J.; Woodcock, C.L.; Peterson, C.L.; Bartholomew, B. Architecture of the SWI/SNF-nucleosome complex. Mol. Cell. Biol. 2008, 28, 6010–6021.
- Lorch, Y.; Maier-Davis, B.; Kornberg, R.D. Mechanism of chromatin remodeling. Proc. Natl. Acad. Sci. USA 2010, 107, 3458–3462.
- Saha, A.; Wittmeyer, J.; Cairns, B.R. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat. Struct. Mol. Biol. 2005, 12, 747–755.
- Kulić, I.; Schiessel, H. Chromatin dynamics: nucleosomes go mobile through twist defects. Phys. Rev. Lett. 2003, 91, 148103.
- Brandani, G.B.; Takada, S. Chromatin remodelers couple inchworm motion with twist-defect formation to slide nucleosomal DNA. PLoS Comput. Biol. 2018, 14, e1006512.
- Fischer, C.J.; Saha, A.; Cairns, B.R. Kinetic model for the ATP-dependent translocation of Saccharomyces cerevisiae RSC along double-stranded DNA. Biochemistry 2007, 46, 12416–12426.
- Li, M.; Xia, X.; Tian, Y.; Jia, Q.; Liu, X.; Lu, Y.; Li, M.; Li, X.; Chen, Z. Mechanism of DNA translocation underlying chromatin remodelling by Snf2. Nature 2019, 567, 409–413
- Yan, L.; Wu, H.; Li, X.; Gao, N.; Chen, Z. Structures of the ISWI–nucleosome complex reveal a conserved mechanism of chromatin remodeling. Nat. Struct. Mol. Biol. 2019, 26, 258–266.
- Li, M.; Xia, X.; Tian, Y.; Jia, Q.; Liu, X.; Lu, Y.; Li, M.; Li, X.; Chen, Z. Mechanism of DNA translocation underlying chromatin remodelling by Snf2. Nature 2019, 567, 409–413.
- Chittori, S.; Hong, J.; Bai, Y.; Subramaniam, S. Structure of the primed state of the ATPase domain of chromatin remodeling factor ISWI bound to the nucleosome. Nucleic Acids Res. 2019, 47, 9400–9409.
- Al-Ani, G.; Malik, S.S.; Eastlund, A.; Briggs, K.; Fischer, C.J. ISWI remodels nucleosomes through a random walk. Biochemistry 2014, 53, 4346–4357.
- Morgan, A.M.; LeGresley, S.E.; Briggs, K.; Al-Ani, G.; Fischer, C.J. Effects of nucleosome stability on remodeler-catalyzed repositioning. Phys. Rev. E 2018, 97, 032422.
- Clapier, C.R.; Cairns, B.R. Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature 2012, 492, 280–284.
- Ludwigsen, J.; Klinker, H.; Mueller-Planitz, F. No need for a power stroke in ISWI-mediated nucleosome sliding. EMBO Rep. 2013, 14, 1092–1097.
- Udugama, M.; Sabri, A.; Bartholomew, B. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol. Cell. Biol. 2011, 31, 662–673.
- Fyodorov, D.V.; Kadonaga, J.T. Dynamics of ATP-dependent chromatin assembly by ACF. Nature 2002, 418, 896–900.
- He, X.; Fan, H.Y.; Narlikar, G.J.; Kingston, R.E. Human ACF1 alters the remodeling strategy of SNF2h. J. Biol. Chem. 2006, 281, 28636–28647.
- Partensky, P.D.; Narlikar, G.J. Chromatin remodelers act globally, sequence positions nucleosomes locally. J. Mol. Biol. 2009, 391, 12–25.
- Rippe, K.; Schrader, A.; Riede, P.; Strohner, R.; Lehmann, E.; Längst, G. DNA sequence-and conformation-directed positioning of nucleosomes by chromatin-remodeling complexes. Proc. Natl. Acad. Sci. USA 2007, 104, 15635–15640.
- Manelyte, L.; Strohner, R.; Gross, T.; Längst, G. Chromatin targeting signals, nucleosome positioning mechanism and non-coding RNA-mediated regulation of the chromatin remodeling complex NoRC. PLoS Genet. 2014, 10, e1004157.
- Winger, J.; Bowman, G.D. The Sequence of Nucleosomal DNA Modulates Sliding by the Chd1 Chromatin Remodeler. J. Mol. Biol. 2017, 429, 808–822.
- Lequieu, J.; Schwartz, D.C.; de Pablo, J.J. In silico evidence for sequence-dependent nucleosome sliding. Proc. Natl. Acad. Sci. USA 2017, 114, E9197–E9205.
- Zhou, C.Y.; Johnson, S.L.; Lee, L.J.; Longhurst, A.D.; Beckwith, S.L.; Johnson, M.J.; Morrison, A.J.; Narlikar, G.J. The yeast INO80 complex operates as a tunable DNA length-sensitive switch to regulate nucleosome sliding. Mol. Cell 2018, 69, 677–688.
- Rippe, K.; Schrader, A.; Riede, P.; Strohner, R.; Lehmann, E.; Längst, G. DNA sequence-and conformation-directed positioning of nucleosomes by chromatin-remodeling complexes. Proc. Natl. Acad. Sci. USA 2007, 104, 15635–15640.
- Manelyte, L.; Strohner, R.; Gross, T.; Längst, G. Chromatin targeting signals, nucleosome positioning mechanism and non-coding RNA-mediated regulation of the chromatin remodeling complex NoRC. PLoS Genet. 2014, 10, e1004157.
- Winger, J.; Bowman, G.D. The Sequence of Nucleosomal DNA Modulates Sliding by the Chd1 Chromatin Remodeler. J. Mol. Biol. 2017, 429, 808–822.
- Lequieu, J.; Schwartz, D.C.; de Pablo, J.J. In silico evidence for sequence-dependent nucleosome sliding. Proc. Natl. Acad. Sci. USA 2017, 114, E9197–E9205.
- Zhou, C.Y.; Johnson, S.L.; Lee, L.J.; Longhurst, A.D.; Beckwith, S.L.; Johnson, M.J.; Morrison, A.J.; Narlikar, G.J. The yeast INO80 complex operates as a tunable DNA length-sensitive switch to regulate nucleosome sliding. Mol. Cell 2018, 69, 677–688.
