Breast cancer is the most common cancer among women worldwide. In the clinic today, imaging techniques like mammography and tissue biopsies are used to diagnose breast cancer. Even though these methods are important in primary diagnosis, they have limitations when it comes to longitudinal monitoring of residual disease after treatment, disease progression, therapy responses, and disease recurrence. Circulating cancer-derived material, including circulating cancer cells (CTCs), circulating DNA (ctDNA), and biomolecules encapsulated in extracellular vesicles, acquired through liquid biopsies are currently focus of research as diagnostic, prognostic, and predictive biomarkers in breast cancer.
- liquid biopsy
- breast cancer
- prediction
- prognosis
- exosomes
- circulating tumor DNA
- circulating tumor cells
1. Introduction
Biomarkers detected through liquid biopsies can potentially aid in early diagnosis, prognostication, and predict response to a specific therapy. Importantly, liquid biopsies can improve longitudinal disease monitoring to detect potential recurrence. The use of liquid biopsies has some advantages over conventional biopsies, such as the ability to capture the intra-tumor heterogeneity between different regions in the same tumor (spatial heterogeneity), as well as between the primary tumor and metastasis in the same patient (temporal heterogeneity)[1][2]. In addition, preservation methods for tissue biopsies can cause biases, an example being formalin fixation causing high levels of C > T/G > A transitions[3]. Liquid biopsies can overcome both preservation and sampling biases, requires shorter collection and analysis time, and is a less invasive method. Despite this, liquid biopsy is not yet implemented as a standard procedure and is used mainly as a complementary test to tissue biopsy.
There are several types of tumor material that can be assessed by liquid biopsies, such as circulating tumor cells (CTCs), cell-free circulating tumor DNA (ctDNA), and extracellular vesicles (EVs) (Figure 1). CTCs represent intact, viable non-hematological cells with malignant features that can be isolated from a blood sample. CTCs are shed from the primary tumors or metastatic lesions into the bloodstream as single cells or clusters. CtDNA is fragmented cell-free DNA (cfDNA) that is released by apoptotic and necrotic tumor cells. Such DNA fragments may harbor cancer-specific mutations that have occurred in the originating cell. In a similar manner, cancer cell-derived EVs carry cargo that reflects the content of the originating cell.
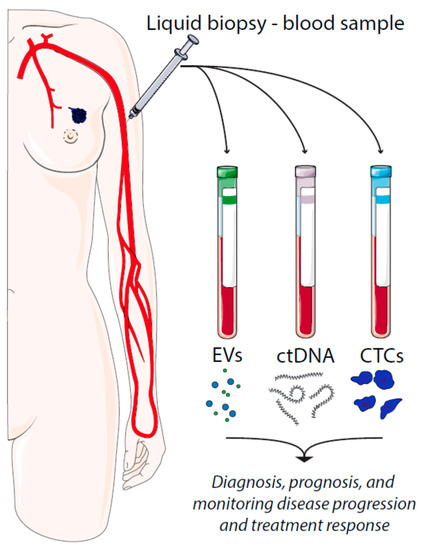
Figure 1. Liquid biopsies in breast cancer and their utilities. EV = extracellular vesicles, ctDNA = circulating tumor DNA, CTCs = circulating tumor cells.
2. Circulating Tumor Cells
Circulating tumor cells (CTCs) are cancer cells that have been shed from the primary tumor or a metastatic lesion into the bloodstream. Even though millions of cancer cells in some primary tumors are shed every day, only a sub-fraction of the CTCs might have the propensity to form distant metastases[4][5][6]. Importantly, in breast cancer, both the number and the characteristics of the CTCs can provide critical information about disease progression and response to therapy[7][8][9][10][11][12]. Therefore, great effort has been put forth to develop protocols enabling detection and characterization of CTCs through liquid biopsies. Molecular characterization of CTCs has provided initial evidence demonstrating that CTCs can serve as surrogates for metastatic disease[13]. Moreover, CTCs isolated from breast cancer patients at early stages can be used as a prognostic tool[14]. Finally, CTCs can be used in real-time monitoring therapy responses at different time points during the disease course and for the detection of relapses[15][16]. Although studying CTCs is a promising approach for better characterizing cancers, there are certain issues inherent to the nature of CTCs that should be considered. CTCs are rare events to the point that it is estimated that there will be only one tumor cell per million blood cells in a sample[17]. Thus, it is vital to have a cancer cell-specific enrichment step before analyzing the features of CTCs. Current approaches for isolation and enrichment include a wide range of commercially available technologies based on either biological properties (cell surface protein expression, viability, invasive capacity) or physical properties (size, density, electric charges, deformability) that distinguish cancer cells from surrounding normal hematopoietic cells. These techniques have recently been reviewed[17][18]. Today, the most frequently used strategy for isolating CTCs is based on capturing cells that express the surface molecule epithelial cell adhesion molecule (EpCAM)[19][20]. EpCAM is considered a common marker protein of epithelial-derived cancers and is involved in several cancer-relevant cellular mechanisms like proliferation and cancer cell stemness[21]. Importantly, EpCAM is not expressed in leukocytes[21]. During the past ten years, there has been an increase in the number of methodologies using EpCAM for the detection of CTCs in the blood of carcinoma patients, such as Magsweeper, MACS, Strep-Tactin, CTC-Chip, and the GO Chip[22]. However, the only method for CTC analyses approved by the U.S. Food and Drug Administration (FDA) is the CellSearch® device[23]. The device was implemented on breast cancer already in 2004 in a study showing that the number of CTCs detected before therapy could predict survival in patients with metastatic breast cancer[11]. Briefly, this method is based on the extraction of 7.5 mL blood into special tubes that are centrifuged to separate solid blood components from plasma. Then, by using ferrofluid nanoparticles coated with EpCAM-targeting antibodies, CTCs are magnetically separated from the bulk of other cells present in the blood. CTCs are then stained with monoclonal antibodies towards cytokeratins (CK18 and CK19) that are specific to epithelial cells. Moreover, CD45 staining is used in order to identify possible contaminating leukocytes remaining in the sample. The cartridge containing stained CTCs is scanned to quantify the tumor cells. In this review we will include the most recent clinical studies that have demonstrated the prognostic value of CTC detection and enumeration in early and metastatic breast cancer using the CellSearch® device (Table 1). Previously in 2016, two other reviews have been published on the same topic[13][24].
Several studies have shown that detection of CTCs at the time of diagnosis (baseline) and at different following-up time points during or after completion of therapy can have a prognostic value for early breast cancer patients. This was recently demonstrated by Trapp et al., who conducted a retrospective study, including node-positive or high-risk node-negative primary invasive breast cancer patients from the SUCCESS A trial (NCT02181101)[25]. The main aim of the study was to evaluate CTCs as a prognostic surveillance-marker during routine follow-up of breast cancer after chemotherapy. They found that the presence of CTCs two years after chemotherapy was associated with decreased disease-free as well as overall survival. Moreover, they reported that the CTC status at the two-year follow-up was an independent prognostic factor of the CTC status at baseline[25]. In another study, Sparano et al. evaluated the prognostic capacity of CTC detection five years after primary diagnosis of lymph-node-positive or high-risk lymph-node-negative breast cancer patients included in the E5103 clinical trial (NCT00433511)[26]. They found that detection of ≥1 CTC in blood from HR+ patients at the five-year follow-up was associated with a higher risk of late recurrence (defined as recurrence five years or later after primary diagnosis)[26]. Similarly, Hall et al. assessed the prognostic value of CTCs identified in non-metastatic breast cancer patients before surgical resection[27] [44]. They found that the presence of ≥1 CTCs before resection was negatively associated with relapse-free and overall survival. This was independent of primary tumor size, grade, and lymph node positivity. Pierga et al. assessed the prognostic and predictive potential of CTCs in data from two phase II multicenter trials (BEVERLY-1 and -2, NCT00820547 and NCT00717405), including patients with non-metastatic inflammatory breast cancer, both HER2+ and HER2−[28]. They found that patients with detectable CTCs at baseline had shorter three-year disease-free survival and overall survival as compared to CTC-negative patients. They also reported that patients that both displayed a pathologic complete response and were negative for CTCs at baseline constituted a subgroup of inflammatory breast cancer patients with particularly high overall survival[28]. Recently, a comprehensive meta-analysis of data from 21 different studies of early-stage non-metastatic breast cancer patients with known CTC baseline counts was reported[29]. The authors concluded that baseline CTC detection was an independent prognostic factor for loco-regional relapse-free intervals, distant disease-free survival, and overall survival, which can be added to well-established prognostic models, including clinicopathological data and pathological complete response status[29]. CTC counts can also predict which patients will benefit from additional radiotherapy after surgery. Goodman et al. studied the effect of radiotherapy on survival of early-stage breast cancer patients that were grouped according to CTC detection at baseline[30]. Whereas radiotherapy after breast-conserving surgery was associated with increased survival within the CTC-positive group, no additional effect of radiotherapy was observed for the CTC-negative group. This indicates that CTC counts could be useful in guiding clinicians in determining which patients will benefit from radiotherapy. CTCs have also been shown to improve the prognosis of metastatic breast cancers. For instance, Larsson et al. evaluated whether detected CTCs and CTC clusters both at baseline and after one, three and six months of systemic therapy, was associated with progression-free survival and overall survival[31]. Although ≥ 5 CTCs and the presence of CTC clusters at baseline indeed were associated with worse prognosis, they found an increased prognostic value of CTC detection over time, suggesting that the dynamic changes in CTC levels are more relevant to prognosis than only baseline enumeration. Moreover, they concluded that the presence of CTC clusters added more significant prognostic value than simple CTC enumeration. Altogether, these results suggest that monitoring CTCs and CTC clusters in blood from metastatic breast cancer patients undergoing systemic therapy may give vital prognostic information about end-point clinical outcomes[31].
As illustrated by the above-mentioned studies, increasing evidence indicates that CTC enumeration can improve prognostication of breast cancer. Other studies have aimed to determine whether receptors on the surface of the CTCs can predict response to therapy. Receptor activator of nuclear factor-kappaB ligand (RANKL) and its receptor RANK play critical roles in breast cancer development, both during initial tumorigenesis and in the formation of secondary tumors in the bone[32]. Today, breast cancer patients with bone metastases are treated with denosumab, an inhibitory antibody targeting RANKL. Recently, Pantano et al. showed that RANK-expressing CTCs were present in blood from breast cancer patients with skeletal metastases[33]. Moreover, the presence of RANK-positive CTCs was associated with an increased response to denosumab treatment. The authors suggested that monitoring RANK-positive CTCs during denosumab treatment might be an important strategy aiding therapy decisions for bone metastatic breast cancers[33]. In HER2+ breast cancers, several researchers have hypothesized that the presence of HER2+ CTCs can predict the response to HER2− directed therapy. Recently, Jaeger et al. reported that HER2 expression in primary tumors and CTCs isolated from patients after surgery was discordant[34]. They hypothesized that this might have important implications for the use of HER2 targeted therapy. Therefore, they suggested that the HER2 status needs to be reassessed after surgery before initiating adjuvant HER2− targeted therapy[34].
In addition to all the reported clinical trials, there are currently many ongoing studies trying to prove the usefulness of CTCs for breast cancer prediction and prognosis. These studies have been recently reviewed by Schochter et al.[35] .
Table 1. Examples of studies demonstrating the clinical relevance of circulating tumor cells and ctDNA.
|
Early Breast Cancer/Advanced Disease |
Liquid Biopsy |
Method |
Prognostic/Predictive Value or Monitoring |
References |
|
Early breast cancer |
CTCs |
CellSearch® |
Prognostic |
|
|
Advanced disease |
CTCs |
CellSearch® |
Prognostic |
[26] |
|
Advanced disease |
CTCs |
CellSearch® |
Prediction |
|
|
Early breast cancer |
CTCs |
CellSearch® |
Prediction |
|
|
Advance disease |
ctDNA |
ddPCR |
Prediction and monitoring |
[36] |
|
Early breast cancer |
ctDNA |
ddPCR |
Prognostic |
[37] |
|
Advanced disease |
ctDNA |
ddPCR |
Prediction and prognostic |
[38] |
|
Advanced disease |
ctDNA |
Signatera™ |
Prognostic |
[39] |
|
Early breast cancer and Locally advanced disease |
ctDNA |
TARDIS |
Prediction and monitoring |
[40] |
3. Cell-Free DNA and Circulating Tumor DNA
DNA is released from both healthy and cancer cells into circulation. Cell-free DNA (cfDNA) found in circulation is highly fragmented and most likely originates from apoptotic and necrotic cells, but the exact origin and mechanism of release are still being investigated[41]. Importantly, primary tumors, circulating tumor cells, and occult and overt metastases release DNA at a higher rate than normal cells, and because circulating tumor DNA (ctDNA) displays mutations characteristic of the progenitor tumor, it can serve as a biomarker for diagnosis, prognosis, and prediction[42]. However, the total amount of ctDNA may represent as low as 0.01% of the total cfDNA. These extremely low concentrations make detection challenging, particularly at the early stages of tumor development as tumor burden is low[43]. Despite these difficulties, researchers have worked out several strategies for the detection of ctDNA in breast cancer. There are two fundamental strategies to analyze ctDNA: (1) Quantification of total cfDNA based on the notion that cancer cells release more DNA than healthy cells, and (2) Specific analysis of ctDNA most often based on available mutation data from tumor tissue.
Once total cfDNA is isolated from the blood, it can be measured by fluorescence-based methods or by real-time quantitative PCR (RT-qPCR)[44]. Tangvarasittichai et al. compared the levels of cfDNA in plasma from breast cancer patients and healthy donors, measured by a fluorometer[45]. They reported significantly higher amounts of plasma cfDNA in stage II, III, and IV breast cancer patients at baseline than in healthy controls. Moreover, tumor size, TNM stage, and metastasis were all significantly correlated with the amount of cfDNA[45]. Importantly, after surgery, the cfDNA level dropped, indicating that cfDNA detection can be used to evaluate the efficacy of surgery. Despite the simplicity of the method and that it could easily be incorporated into clinical practice, the diagnostic value of cfDNA quantitation is limited due to the significant overlap of cfDNA concentration in normal and cancer patients and the low ability to distinguish early breast cancer from healthy patients[46].
Measuring the level of ctDNA in cancer patients is a more specific method than measuring cfDNA, and for this reason, it could have a greater potential as a novel method for oncological applications. ctDNA fluctuations during the course of the disease could be correlated with clinical outcome. It was demonstrated by Kruger et al. that high ctDNA molecule numbers and at least two breast cancer-specific mutations, measured by next-generation sequencing (NGS), correlate with poor clinical outcome in ER-positive, HER2-negative metastatic breast cancer patients[47]. Over the last decade, several methodologies have been developed as new viable tools for genomic screening of ctDNA, allowing early detection and management of breast cancer. There are two different approaches for the genomic analysis of ctDNA: (1) Targeted approaches in which a single or few tumor-specific mutations known are used for monitoring residual disease and (2) Untargeted screening that aims at genome-wide analysis for copy number aberrations or point mutations by whole-genome sequencing (WGS) or whole-exome sequencing (WES)[48]. The main disadvantage of targeted strategies is that detailed information about the tumor genome is required. Targeted monitoring can, however, be extremely sensitive, as mutations can be detected at an allele frequency down to 0.01%, with high specificity and at fast and cost-effective rates[49]. Advantages of untargeted strategies include its ability to identify novel changes occurring during tumor treatment and the fact that prior information about the primary tumor’s genome is not required. However, high concentrations of ctDNA are required for reliable reconstruction of tumor-specific genome-wide changes, and usually, they show lower sensitivity (5–10%)[50]. In the following paragraphs different targeted strategies will be reviewed, pointing out those that have been used in breast cancer studies (Table 1).
Detection and quantification of predetermined alleles from ctDNA by Sanger sequencing, single and multi-locus PCRs, and RT-qPCR have established methods in clinical oncology[51][52]. However, these methods are limited to detecting mutant allele frequencies (MAFs) well above 1%, which would leave many cancers smaller than 10 cm3 undetected because the expected mutation-carrying ctDNA concentration in plasma for these patients can be 0.1% or lower of total cfDNA[53][54]. Advances in digital PCR, such as digital droplet PCR (ddPCR) and the BEAMing technology, allow absolute quantification of allele frequencies as low as 0.01%[48]. Both technologies have been shown to be able to detect PIK3CA mutations in plasma ctDNA from breast cancer patients[36][55]. PIK3CA encodes the catalytic p110a subunit of PI3-kinase and is mutated in 30–40% of all breast cancer[56]. By using ddPCR, Kodahl et al. recently showed that in breast cancer patients with advanced disease, there is a strong concordance between PIK3CA mutations in metastatic tumor tissue and serum ctDNA[57]. Moreover, although including only a limited number of samples, they observed that the level of PIK3CA mutated ctDNA dropped in patients that displayed response to therapy. This suggests that ddPCR-mediated detection of ctDNA PIK3CA mutations can be used in early monitoring of therapy response[36]. Several studies have indeed shown that ctDNA PIK3CA mutations can be detected in metastatic breast cancer patients[58][59][60]. In contrast, Beaver et al. focused their analyses on blood samples from 15 early-stage breast cancer patients that harbored PIK3CA mutations in the primary tumors[37]. In 14 patients (93%), concordant PIK3CA mutations were detected in ctDNA in blood samples taken before surgery. CtDNA PIK3CA mutational status was also analyzed after surgery for some of the patients, and 5 out of 10 still had detectable ctDNA PIK3CA mutations. However, further studies have to be undertaken to assess whether detected PIK3CA mutated ctDNA after surgery is an identifier of patients with a higher risk of recurrence[37]. Interestingly, a recent device detecting PIK3CA mutations on ctDNA has been approved by the FDA. The therascreen PIK3CA RGQ PCR Kit is a laboratory test able to detect 11 mutations in the PIK3CA gene from patients with advanced or metastatic breast cancer. The presence of PIK3CA mutations is linked with response to treatment with PIQRAY® (alpelisib)[61]. Thus this laboratory test may help doctors identify breast cancer patients who should be treated with PIQRAY®. Another attractive target for ctDNA mutational analyses is the ESR1 gene encoding estrogen receptor alpha (ERα ESR1 mutations are associated with resistance to aromatase inhibitors (AIs) that are used as estrogen-deprivation therapy in postmenopausal women[62][63]. Importantly, several studies have shown that the ESR1 mutations can be selected during AI treatment of metastatic HR+ breast cancers, which can be detected in ctDNA[37][64][65]. In a comprehensive study, Chandarlapaty et al. performed allele-specific ddPCR of ESR1 mutations Y537S and D538G, which lead to ligand-independent constitutive activation of ERα in blood samples from 541 ER+/HER2− breast cancer patients with metastatic diseases that all had undergone AI therapy in different settings (BOLERO-2, NCT00863655)[37]. They detected ESR1 Y537S and/or D538G mutations in ctDNA in 28.8% of the patients. Importantly, the presence of the ESR1 ctDNA mutations was associated with reduced overall survival. Thus, monitoring ESR1 ctDNA mutations can be important in disease control of metastatic ER+ cancers and potentially give valuable information on which patients will benefit from hormonal therapy.
The above-mentioned studies indicate that digital PCR is a sensitive method that is suited for detecting allele variation in ctDNA from clinical samples. A recent study assessed the agreement between the BEAMing and ddPCR techniques in detecting PIK3CA and ESR1 ctDNA mutations plasma samples from patients with HR+, HER2− metastatic breast cancer enrolled in phase 3 PALOMA-3 trial (NCT01942135)[66]. The authors concluded that the concordance between the techniques is very good, suggesting their sufficient reliability and reproducibility for clinical applications. Of note, these two technologies have been combined in the clinically validated OncoBEAM™ device, which has been tested in several clinical studies, including breast cancer[55][67][68].
The revolution within massively parallel NGS technologies has opened up a new era within personalized medicine. The sensitivity of these methods enables the detection of patient-specific chromosomal structural aberrations, copy number variations, and single nucleotide polymorphisms (SNPs) in both tissue samples and liquid biopsies. Both unbiased WGS and more targeted approaches may be used. Even though proof-of-concept studies have been reported, ctDNA analyses by WGS approaches appear to be somewhat limited by the low abundance of ctDNA fraction in total cfDNA, short ctDNA fragment lengths, as well as by high error rates in the detection of rare allele variants[69][70]. Therefore, targeted approaches are being developed for ctDNA analyses aimed at specifically detecting cancer-specific mutations that have been identified in the primary tumor. Currently, there are several targeted NGS experimental procedures validated in breast cancer patient ctDNA samples. These will be discussed below.
In 2019, the first custom-built NGS-based ctDNA test, Signatera™, was launched (https://www.natera.com/oncology/signatera-advanced-cancer-detection). The aim of this technology is to improve the detection of minimal residual disease after surgery as well as earlier detection of disease recurrence. Signatera™ provides each individual patient with a customized blood test tailored to match the clonal mutations found in the individual’s tumor tissue by WES. Coombes et al. validated the clinical performance of Signatera™ on breast cancer patient samples[38]. They performed upfront WES of tumor tissue from 49 patients and subsequently designed personalized panels of 16 SNPs or indels that were targeted by multiplex sequencing of plasma ctDNA. Importantly, this method was able to detect disease relapse across different subtypes of breast cancer up to two years earlier than imaging in patients with early-stage breast cancer. Other clinical studies in colon cancer[71], lung cancer[72], and bladder cancer[73] support the use of this approach. Recently, it was reported that targeted digital sequencing (TARDIS) of ctDNA might be a highly sensitive method to predict a pathological complete response to neoadjuvant treatment in early and locally advanced breast cancers[39]. Following the WES of tumor biopsies and matched germline samples of 33 patients, the authors analyzed plasma samples for personal mutations (ranging from 6–115 per person) before neoadjuvant treatment. By employing this sensitive strategy, they detected ctDNA in 100% of the samples. They then monitored ctDNA at different time points in 22 patients that were subjected to neoadjuvant treatment and found that ctDNA levels were higher in patients with the residual disease than those that displayed a pathological complete response. The authors argue that this is a promising test to identify women in whom tumors are effectively eliminated by the pre-surgery treatment and, therefore, may not get any additional benefits from undergoing surgery.
ctDNA is believed to be shed across the entire tumor and by all tumor sites, including metastases; thus, it may be a useful tool for addressing tumor heterogeneity. In a proof-of-principle study, De Mattos-Arruda et al. subjected primary tumor, lung metastasis, and plasma samples from a metastatic ER+/HER2− breast cancer patient to massively parallel sequencing targeting 300 genes that are frequently mutated in cancer[74]. All mutations identified in the primary tumor and metastatic lesion were captured in the ctDNA. Importantly, the mutant allele fractions identified in ctDNA varied over time and reflected the clinical response to targeted therapy. Likewise, Murtaza et al. performed a comprehensive case study of an ER+/HER2+ metastatic breast cancer patient with multiple metastatic lesions[74]. They conducted exome and targeted deep amplicon sequencing on samples taken from the primary tumor, several metastatic sites, and serial samples of blood taken throughout the course of the disease. Importantly, ctDNA captured ubiquitous stem mutations (detected in all tumor samples), metastatic-clade mutations (detected in metastatic lesions), and private mutations (only detected in one specific tumor sample). Moreover, disease progression at specific metastatic lesions was reflected in changes in circulating levels of specific private somatic mutations. This indicates that intratumor heterogeneity and clonal evolution can be monitored by serial analyses of ctDNA, which may improve disease control and guide therapy decisions.
4. Circulating Exosomal RNA Species and Proteins
Hence, far, we have focused our discussion on CTCs and ctDNA as potential biomarkers in breast cancer. These have by far been the most extensively studied analytes in clinical studies due to the implementation of devices like CellSearch®, and the recent innovation within technologies for mutational analyses like ddPCR, WES/WGS, and targeted deep sequencing. However, other classes of tumor-derived molecules such as proteins and RNA-species that are released into circulation may potentially provide vital information about cancer development and progression. In this chapter, we will review literature describing studies suggesting that circulating exosomal RNA and proteins detected through liquid biopsies may potentially be implemented as diagnostic, prognostic, or predictive biomarkers (Table 2).
All cells produce and release membranous vesicles into the extracellular environment. Extracellular vesicles (EVs) are generally heterogeneous but can broadly be divided into two classes based on size and mechanisms of biogenesis[75]. These are microvesicles (also referred to as ectosomes) of 100–1000 nm that are formed by plasma membrane shedding, and exosomes of 30–150 nm that derive from the endocytic pathway and are released when multivesicular bodies (MVBs) fuse with the plasma membrane[75][76]. Exosomes, in particular, have gained substantial attention over the last few years as experimental evidence has demonstrated that they contribute to the development and progression of human diseases, including cancer[75][77][78]. It has been demonstrated that cancer cells release more exosomes compared to normal cells as a response to pathophysiological conditions like hypoxia and low pH in the tumor microenvironment or to oncogenic activities within the cells[79][80][81]. Exosomes carry biomolecular cargo, including proteins, lipids, DNA, and RNA species that reflect the content of their originating cells[75][82]. They play key roles in intercellular communication and can deliver their content to neighboring cells in a paracrine fashion. Importantly, exosomes are also detected in body fluids, including blood, suggesting that they can act as mediators of long-distance cellular signaling[82][83][84]. In cancer, exosomes may promote tumor cell migration, invasion, and formation of distant metastatic niches[75][77][81]. They have also been shown to play a role in cancer cell immune evasion[85]. As exosomes are present in body fluids and provide a protective compartment for their biological content, they are promising sources for biomarkers that can be detected through liquid biopsies[82][83][84]. The field still faces challenges in terms of standardization of exosome isolation protocols and lack of proper normalization strategies. This has hampered clinical implementation, and the number of registered clinical studies on cancer exosomes is still scarce. However, several published studies have indeed suggested that analyses of exosomes from body fluids can aid early diagnosis and disease progression monitoring of breast cancer, as well as other cancers[82].
Exosomes isolated from breast cancer patients have been shown to have distinct protein- and RNA contents compared to exosomes derived from healthy donors[86][87]. In this regard, exosomal microRNAs (miRNAs) have been the focus of research in several studies. Many miRNAs are incorporated into exosomes and transferred to other cells where they directly regulate target mRNAs[88]. Compared to larger RNA molecules, miRNAs are remarkably stable, which aid their detection in circulating exosomes. Hannafon et al. performed comprehensive RNA-sequencing-based profiling of exosomal miRNAs derived from cultivated breast cancer cell lines[89]. From their list of exosomal miRNAs, they showed that the levels of miR-1246 and miR-21 were significantly higher in plasma exosomes from breast cancer patients than from healthy donors. Blood samples from triple-negative breast cancer patients have been shown to contain increased levels of exosomal miR-373 compared to other breast cancer subtypes, indicating that it is a potential biomarker for diagnosis of basal-like breast cancers[90]. Stevic et al. analyzed the presence of a panel of 45 exosomal miRNAs in plasma from 435 patients diagnosed with either HER2+ or triple-negative breast cancer and indeed confirmed that the exosomal miRNA content differed among the two breast cancer subtypes[91]. Moreover, they reported that specific exosomal miRNAs are associated with clinicopathological parameters, including tumor size, lymph node status, and pathological complete response to neoadjuvant therapy. Recently, it was shown that circulating exosomal miR-21 levels are significantly higher in breast cancer patients with distant metastases compared to those that only had local disease[92]. In HER2+ cancers, the level of miR-21 dropped after treatment with trastuzumab in a neoadjuvant setting, indicating that miR-21 levels could be used in monitoring the treatment response of HER2+ cancers[92]. Sueta et al. compared the expression of 384 exosomal miRNAs in serum from breast cancer patients with recurrence and patients without recurrence[93]. They identified 11 miRNAs that were differentially expressed in the two groups of patients, suggesting that they could be used as prognostic biomarkers. Exosomal miRNAs can also distinguish patients with invasive ductal carcinoma (IDC) from patients diagnosed with ductal carcinoma in situ (DCIS). This is exemplified by miR-223-3p levels that were significantly higher in patients with IDC compared to patients with DCIS or healthy individuals[94]. Importantly, miR-223-3p levels could also predict which DCIS samples ultimately were upgraded to IDC at final pathologic diagnosis. All the above-mentioned studies have been done in exosomes isolated from blood samples. Recently, however, Hirschfeld and coworkers analyzed the expression of 13 miRNAs from exosomes isolated from urine samples from breast cancer patients and healthy volunteers[95]. They suggested that a panel of four urine exosomal miRNAs, including miR-424, miR-423, miR-660, and let7-i, could be used as biomarkers for breast cancer detection.
Even though the majority of studies on circulating exosomal RNA molecules have been focused on miRNAs, a few studies have shown that exosomal mRNAs and long noncoding RNAs (lncRNAs) might be used as biomarkers. Rodriguez et al. showed that a set of metastasis- and stemness-related mRNAs were higher expressed in plasma exosomes from breast cancer patients with poor disease outcomes than in those that were isolated from patients with good outcomes[96]. Increased circulating exosomal mRNA expression of the drug metabolic enzyme glutathione S-transferase P1 (GSTP1) was associated with drug resistance in breast cancer patients[97]. In HR+ breast cancer, elevated circulating exosomal levels of mRNAs encoding cell cycle-regulated thymidine kinase 1 (TK1) and cyclin-dependent kinase 9 (CDK9) were associated with poor clinical response to the CDK4/CDK6 inhibitor palbociclib[98]. Recently, it was reported that higher levels of the lncRNA HOTAIR were found in circulating exosomes in breast cancer patients than in healthy individuals[99]. Exosomal HOTAIR levels were also associated with poor prognosis, and it was suggested as a possible predictor of response to chemotherapy and tamoxifen treatment. Zhong et al. reported that serum exosomal levels of the lncRNA H19 were significantly higher in breast cancer patients than healthy controls[100]. Finally, in HER2+ breast cancers, elevated serum exosomal levels of the lncRNA SNHG14 were associated with resistance to trastuzumab[101].
The protein content of exosomes reflects their mode of biosynthesis, and they are typically enriched with endosomal/lysosomal proteins like tetraspanins (CD9, CD63, CD81, and CD82), ESCRT complex proteins (e.g., ALIX and TSG101), Rab proteins, Syndecan-1, and Syntenin-1 [75][76]. Quantification of exosomal CD82 in serum has indeed been suggested as a strategy for early diagnosis of breast cancer[102]. Heat shock proteins (Hsps), including Hsp90 and Hsp70, are also frequently found in exosomes[103]. Hsps are often upregulated in cancer due to proteotoxic stress[104], and recently, a prospective clinical pilot study reported that plasma from both breast cancer and non-small cell lung cancer patients contained increased levels of exosomal Hsp70 compared to healthy donors[105]. Furthermore, exosomal Hsp70 levels were significantly higher in patients with metastatic disease compared to those with non-metastatic disease. Finally, plasma exosomal Hsp70 levels could predict the response to therapy. Breast cancer patients have also been shown to have elevated levels of circulating exosomal fibronectin, developmental endothelial locus-1 (Del-1), and the 2B splice variant of survivin (survivin-2B), compared to healthy controls[106][107][108][109]. In a study including 32 breast cancer samples, 75% of the patients were found to have increased levels of circulating exosomal glypican-1 (GPC1), a heparan sulfate proteoglycan that is overexpressed in breast cancer, compared to a control group[110]. Exosomes isolated from pleural effusions from breast cancer patients have increased levels of the epithelial marker proteins EpCAM and CD24 compared to controls [128]. Whereas CD24 also was detected in serum exosomes of breast cancer patients, conflicting results have been reported for exosomal EpCAM in blood. Whereas Rupp et al. failed to detect EpCAM on the surface of circulating exosomes in serum possibly due to its proteolytic cleavage by serum proteases, Fang and coworkers were able to isolate and detect increased levels of EpCAM-positive exosomes from plasma from breast cancer patients, using a microfluidic chip device[85] [111][112]. A comprehensive phosphoproteomic study of plasma exosomes indicated that breast cancer could be diagnosed based on cancer-specific phosphorylation events that are reflected in the phosphoprotein content of circulating exosomes[113]. Three exosomal phosphoproteins, RALGAPA2, PRKG1, and TJP2, were validated and suggested as potential breast cancer biomarkers.
Several reports have shown that the proteome of breast cancer-derived exosomes reflects the subtype of the malignant cells. For instance, it has been shown that ectopic overexpression of the HER2− encoding ERBB2 gene in the immortalized human mammary luminal epithelial cell line HB4a altered the proteomic landscape of the secreted exosomes[114]. Moreover, the overexpressed HER2 receptor was packed into and could be detected in exosomes. Consistency between the HER2 expression status in the breast cancer tissue and circulating exosomes have been demonstrated[112][115][116]. Finally, Rontogianni et al. showed that triple-negative breast cancers could be distinguished from HER2+ breast cancers by proteomic profiling of circulating exosomes[116].
The protein content of circulating exosomes in breast cancer patients can potentially predict the response to therapy. It has been suggested that the HER2 receptor on exosomes might act as a decoy for trastuzumab, a HER2-targeting monoclonal antibody used in the clinic today[115]. By doing this, exosomal HER2 may directly counteract the efficacy of trastuzumab. Ning et al. showed that circulating exosomal levels of Ubiquitin carboxyl-terminal hydrolase-L1 (UCH-L1), which may upregulate drug resistance-associated P-glycoprotein (P-gp) levels, can predict the response to chemotherapy in breast cancer patients[117]. Similar observations were done for another P-gp regulator, Transient receptor potential channel 5 (TrpC5), that were detected in circulating exosomes from patients that had undergone chemotherapy and found it to be a predictor of acquired chemoresistance in metastatic breast cancer[118][119].
Table 2. Examples of studies suggesting the potential of circulating exosomal RNA and proteins as liquid biopsies in breast cancer.
|
Exosomal Content |
Prognostic/Predictive Value/ Monitoring/Diagnostic |
References |
|
miRNA |
Diagnosis |
|
|
miRNA |
Prognosis |
|
|
miRNA |
Prediction |
[91] |
|
miRNA |
Monitoring |
[92] |
|
mRNA/lncRNA |
Diagnosis |
|
|
mRNA/lncRNA |
Prognosis |
|
|
mRNA/lncRNA |
Prediction |
[101] |
|
Protein |
Diagnosis |
|
|
Protein |
Prediction |
3. Discussion and future perspectives
In order to have clinical utility, cancer biomarkers should fulfill a set of analytical and clinical criteria[120][121]. High analytical performance of a biomarker, measured as specificity, precision, accuracy, sensitivity, and good linear range, does not necessarily directly imply its clinical validity or utility. The clinical validity of a biomarker is the ability of the biomarker to accurately identify patients with the targeted pathological state, while the utility measures the benefit (such as reduced mortality) of using the biomarker in clinical settings[120][121]. During the last few years, there has been a tremendous progression in the development of sensitive technologies allowing the detection and analyses of tumor materials in body fluids. Cancer-derived cells and molecules can be isolated through liquid biopsies, and several studies have demonstrated that they can give vital information about the diagnosis, disease progression, and therapy response in breast cancer as well as other cancers. Although being promising, the implementation of such biomarkers into the clinical practice requires a better understanding of underlying shedding/releasing mechanisms, as well as dynamics and kinetics of the circulating tumor material. Moreover, there are still important technical issues that need to be resolved to improve reproducibility[122]. Therefore, in spite of the huge e ort put in by the scientific community, the majority of liquid biopsy assays still lack evidence of clinical validity and, in particular, clinical utility. Therefore, their use is still mostly confined to research purposes and clinical studies[123].
CTCs might give important information about tumor burden and might also contain molecules reflecting cancer-specific features. An important limitation of CTCs is that they may not fully reflect the biology of the underlying tumor. In addition, cancer cells are not confined to one phenotype and might have epithelial, mesenchymal, or stem-like characteristics[124]. Currently, the CellSearch® platform is the only one licensed by the FDA for the isolation of CTCs and their prognostic enumeration in breast cancer[23]. However, the standard CellSearch® platform enriches CTCs based on the expression of the epithelial marker protein EpCAM. In breast cancer, as well as other cancers, several lines of evidence suggest that the most aggressive tumor cells are those that have undergone epithelial-mesenchymal transition (EMT), a process that is associated with downregulation of EpCAM as well as other adhesion molecules[125][126]. EMT is associated with increased propensity to metastasize, acquired stem cell-like features, and drug resistance[127]. Thus, only focusing on EpCAM-expressing cells might confer serious limitations to the studies, leaving groups of CTCs undetected[128]. Another concern is that some studies use a cut of only 1 CTC per 7.5 mL of blood to score a sample as CTC-positive. This indicates that the assay might be prone to inter-laboratory variations in image interpretation, which might impact its robustness. Moreover, rare EpCAM-positive “CTC-like” cells can be detected in healthy women, which might preclude the interpretation of the test[129]. Finally, different types of tumors have different propensity to shed cells. As breast tumors generally display a high degree of heterogeneity, certain subpopulations of cells might more easily be shed into the bloodstream than others[130]. This means that the CTCs detected in liquid biopsies might not reflect the actual intratumoral heterogeneity of a breast tumor. In spite of this, several studies have shown that CTCs reflect the tumor burden, and CellSearch®-based CTC detection both at baseline and during therapy might give vital information about residual disease and recurrence. However, further standardization of the assays addressing all the above-mentioned limitations need to be addressed before CTC enumeration can implemented in the clinic as an independent assay to guide treatment decisions[71][72].
Detection of ctDNA that reflects the mutational status and clonal evolution of a tumor holds great promise for future monitoring of cancer progression and drug response in breast cancer. The development of high-sensitivity technologies enabling detection of mutant allele frequencies of <0.01% has, without doubt, revolutionized this field[21][47]. However, ctDNA constitutes only a minor fraction of total cfDNA, and reliable and reproducible detection of low allele frequency variants
is still challenging[131]. This might, for instance, hamper the detection of metastatic site-specific private mutations that have occurred during clonal evolution. Moreover, it is important to distinguish ctDNA mutations from randomly mutated cfDNA derived from noncancerous blood cells[130]. Another obvious point is that, as for CTCs, not all tumors shed DNA[130]. As mentioned above, a good concordance was demonstrated between ddPCR and BEAMing techniques in detecting PIK3CA and ESR1 ctDNA mutations in plasma samples from breast cancer patients[66]. In contrast, a recent study reported a significant discordance between four commercial NGS-based ctDNA tests, particularly in the detection of allele frequency variants <1%[132]. Therefore, before the implementation of ctDNA tests in the clinic as independent diagnostic, prognostic, or predictive assays, standardization in terms of how the samples are obtained and how the analyses are performed are needed. However, the prospect of using ctDNA analyses for early detection of disease recurrence is promising and might complement conventional biopsies.
It is generally accepted that secreted exosomes provide a protective compartment for circulating biomolecules. Furthermore, in breast cancer and other cancers, tumor-derived exosomes may contain cargo that can give vital information about disease development and progression. However, as illustrated by the limited number of comprehensive clinical studies on exosomes and breast cancer, the field faces challenges that need to be resolved before being clinically applicable. First of all, further research should be conducted on the selection and packaging mechanisms of cargo in the originating cancer cell to better evaluate whether the exosome content in a given sample is based on specific selection or random incorporation of the secreted cargo. In the latter case, inter- and intraindividual heterogeneity of exosomal content might preclude their implementation as reliable biomarkers. Moreover, sensitive methods for purification of exosomes that excludes contaminants like lipoprotein particles, microvesicles, and protein complexes, are needed[133]. Such methods should be optimized based on the nature of the biomolecules that are going to be analyzed. Multiparametric analyses of different exosomal biomolecules might thus require several samples. Finally, the protocols need to be fast and convenient to be used in clinical practice. Today, the gold-standard method for exosome isolation is ultracentrifugation, which is a tedious process that might require large sample volumes. Nevertheless, with efforts being put in developing new isolation protocols like those based on microfluidics and establishing standards applied to all steps in sampling, purification, and subsequent analyses, circulating exosomal-derived biomarkers might provide additive information refining diagnosis and aiding disease control in breast cancer[134].
Most studies on liquid biopsies have been focused on a single analyte (CTCs, ctDNA, exosomes, etc.). This is about to change, and more initiatives are now being taken to develop multiparametric assays that incorporate data from separate platforms analyzing different analytes. Such combinatorial strategies require powerful statistical tools and algorithms that can integrate and standardize data obtained from different platforms. In this regard, PROLIPSY (NCT04556916) is a multinational effort, which is exploring whether a combination of CTCs, ctDNA, and tumor-secreted exosomes might assist in early detection and prognostic profiling of prostate cancer. CancerSEEK is an already commercialized blood-based assay that looks at ctDNA and protein biomarkers in parallel. Cohen et al. conducted a multicenter study, enrolling 1000 patients bearing different types of cancer. Their results showed that they could achieve early-detection rates of 69–98% for five different malignancies (ovary, liver, stomach, pancreas, and esophagus cancer), with a false-positive rate of less than 1%. Regarding breast cancer, this test could only detect around 30% of the cases, so further improvements are needed[57]. Finally, another example is the study conducted by Ye, Z. et al., where they investigate individual and joint e ects of CTC and cfDNA on clinical outcomes of metastatic breast cancer (MBC) patients[135]. The main conclusion was that CTC and total cfDNA levels were individually and jointly associated with progression-free survival and overall survival in metastatic breast cancer patients[135].
4. Conclusions
Currently, exhaustive validation in clinical trials is required to boost the clinical utility of liquid biopsies. More studies are needed to improve the test’s accuracy and sensitivity of the tests. Moreover, additional investigations are required to clarify whether liquid biopsy provides a representative sampling of all genetic clones within a tumor or if there is a bias to specific subregions of the tumor.
This entry is adapted from the peer-reviewed paper 10.3390/ijms21249457
References
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892.
- Beca, F.; Polyak, K. Intratumor Heterogeneity in Breast Cancer. Adv. Exp. Med. Biol. 2016, 882, 169–189.
- Hongdo Do; Alexander Dobrovic; Sequence Artifacts in DNA from Formalin-Fixed Tissues: Causes and Strategies for Minimization. Clinical Chemistry 2015, 61, 64-71, 10.1373/clinchem.2014.223040.
- Tellez-Gabriel, M.; Brown, H.K.; Young, R.; Heymann, M.F.; Heymann, D. The Challenges of Detecting Circulating Tumor Cells in Sarcoma. Front. Oncol. 2016, 6, 202.
- Liotta, L.A.; Saidel, M.G.; Kleinerman, J. The significance of hematogenous tumor cell clumps in the metastatic process. Cancer Res. 1976, 36, 889–894.
- Butler, T.P.; Gullino, P.M. Quantitation of cell shedding into efferent blood of mammary adenocarcinoma. Cancer Res. 1975, 35, 512–516.
- Magbanua, M.J.M.; Rugo, H.S.; Wolf, D.M.; Hauranieh, L.; Roy, R.; Pendyala, P.; Sosa, E.V.; Scott, J.H.; Lee, J.S.; Pitcher, B.; et al. Expanded Genomic Profiling of Circulating Tumor Cells in Metastatic Breast Cancer Patients to Assess Biomarker Status and Biology Over Time (CALGB 40502 and CALGB 40503, Alliance). Clin. Cancer Res. 2018, 24, 1486–1499.
- Strati, A.; Nikolaou, M.; Georgoulias, V.; Lianidou, E.S. Prognostic Significance of TWIST1, CD24, CD44, and ALDH1 Transcript Quantification in EpCAM-Positive Circulating Tumor Cells from Early Stage Breast Cancer Patients. Cells 2019, 8, 652.
- Hayes, D.F.; Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Miller, M.C.; Matera, J.; Allard, W.J.; Doyle, G.V.; Terstappen, L.W. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 2006, 12, 4218–4224.
- Cristofanilli, M.; Hayes, D.F.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Reuben, J.M.; Doyle, G.V.; Matera, J.; Allard, W.J.; Miller, M.C.; et al. Circulating tumor cells: A novel prognostic factor for newly diagnosed metastatic breast cancer. J. Clin. Oncol. 2005, 23, 1420–1430.
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791.
- Pierga, J.Y.; Bidard, F.C.; Mathiot, C.; Brain, E.; Delaloge, S.; Giachetti, S.; de Cremoux, P.; Salmon, R.; Vincent-Salomon, A.; Marty, M. Circulating tumor cell detection predicts early metastatic relapse after neoadjuvant chemotherapy in large operable and locally advanced breast cancer in a phase II randomized trial. Clin. Cancer Res. 2008, 14, 7004–7010.
- Bidard, F.C.; Proudhon, C.; Pierga, J.Y. Circulating tumor cells in breast cancer. Mol. Oncol. 2016, 10, 418–430.
- Martos, T.; Casadevall, D.; Albanell, J. Circulating Tumor Cells: Applications for Early Breast Cancer. Adv. Exp. Med. Biol. 2020, 1220, 135–146.
- Pachmann, K.; Schuster, S. The Value of Monitoring the Behavior of Circulating Tumor Cells at the End of Endocrine Therapy in Breast Cancer Patients. Cancers (Basel) 2018, 10, 407.
- Medford, A.J.; Dubash, T.D.; Juric, D.; Spring, L.; Niemierko, A.; Vidula, N.; Peppercorn, J.; Isakoff, S.; Reeves, B.A.; LiCausi, J.A.; et al. Blood-based monitoring identifies acquired and targetable driver HER2 mutations in endocrine-resistant metastatic breast cancer. NPJ Precis. Oncol. 2019, 3, 18.
- Marta Tellez Gabriel; Lidia Rodriguez Calleja; Antoine Chalopin; Benjamin Ory; Dominique Heymann; Circulating Tumor Cells: A Review of Non–EpCAM-Based Approaches for Cell Enrichment and Isolation. Clinical Chemistry 2016, 62, 571-581, 10.1373/clinchem.2015.249706.
- Sandhya Sharma; Rachel Zhuang; Marisa Long; Mirjana Pavlovic; Yunqing Kang; Azhar Ilyas; Waseem Asghar; Circulating tumor cell isolation, culture, and downstream molecular analysis. Biotechnology Advances 2018, 36, 1063-1078, 10.1016/j.biotechadv.2018.03.007.
- Habli, Z.; AlChamaa, W.; Saab, R.; Kadara, H.; Khraiche, M.L. Circulating Tumor Cell Detection Technologies and Clinical Utility: Challenges and Opportunities. Cancers (Basel) 2020, 12, 930.
- Racila, E.; Euhus, D.; Weiss, A.J.; Rao, C.; McConnell, J.; Terstappen, L.W.; Uhr, J.W. Detection and characterization of carcinoma cells in the blood. Proc. Natl. Acad. Sci. USA 1998, 95, 4589–4594.
- Laura Keller; Stefan Werner; Klaus Pantel; Biology and clinical relevance of EpCAM. Cell Stress 2019, 3, 165-180, 10.15698/cst2019.06.188.
- Petra Bankó; Sun Young Lee; Viola Nagygyörgy; Miklós Zrínyi; Chang Hoon Chae; Dong Hyu Cho; András Telekes; Technologies for circulating tumor cell separation from whole blood. Journal of Hematology & Oncology 2019, 12, 1-20, 10.1186/s13045-019-0735-4.
- Lori M. Millner; Mark W. Linder; Roland Valdes; Circulating Tumor Cells: A Review of Present Methods and the Need to Identify Heterogeneous Phenotypes. Annals of clinical and laboratory science 2013, 43, 295-304, .
- Jin Sun Lee; Mark Jesus M. Magbanua; John W. Park; Circulating tumor cells in breast cancer: applications in personalized medicine. Breast Cancer Research and Treatment 2016, 160, 411-424, 10.1007/s10549-016-4014-6.
- Elisabeth Trapp; Wolfgang Janni; Christian Schindlbeck; Julia Jückstock; Ulrich Andergassen; Amelie De Gregorio; Marianna Alunni-Fabbroni; Marie Tzschaschel; Arkadius Polasik; Julian G Koch; et al. Presence of Circulating Tumor Cells in High-Risk Early Breast Cancer During Follow-Up and Prognosis. JNCI Journal of the National Cancer Institute 2018, 111, 380-387, 10.1093/jnci/djy152.
- Joseph A. Sparano; Anne O’Neill; Katherine Alpaugh; Antonio C. Wolff; Donald W. Northfelt; Chau T. Dang; George W. Sledge; Kathy D. Miller; Association of Circulating Tumor Cells With Late Recurrence of Estrogen Receptor–Positive Breast Cancer. JAMA Oncology 2018, 4, 1700-1706, 10.1001/jamaoncol.2018.2574.
- Carolyn S. Hall; Mandar G. Karhade; Jessica B. Bowman Bauldry; Lily M. Valad; Henry M. Kuerer; Sarah M. DeSnyder; Anthony Lucci; Prognostic Value of Circulating Tumor Cells Identified Before Surgical Resection in Nonmetastatic Breast Cancer Patients.. Journal of the American College of Surgeons 2016, 223, 20-29, 10.1016/j.jamcollsurg.2016.02.021.
- Jean-Yves Pierga; F.-C. Bidard; A. Autret; T. Petit; F. Andre; F. Dalenc; C. Levy; Jean-Marc Ferrero; G. Romieu; J. Bonneterre; et al. Circulating tumour cells and pathological complete response: independent prognostic factors in inflammatory breast cancer in a pooled analysis of two multicentre phase II trials (BEVERLY-1 and -2) of neoadjuvant chemotherapy combined with bevacizumab. Annals of Oncology 2017, 28, 103-109, 10.1093/annonc/mdw535.
- François-Clément Bidard; Stefan Michiels; Sabine Riethdorf; Volkmar Mueller; Laura J. Esserman; Anthony Lucci; Bjørn Naume; Jun Horiguchi; Rafael Gisbert-Criado; Stefan Sleijfer; et al. Circulating Tumor Cells in Breast Cancer Patients Treated by Neoadjuvant Chemotherapy: A Meta-analysis. JNCI: Journal of the National Cancer Institute 2018, 110, 560-567, 10.1093/jnci/djy018.
- Chelain R. Goodman; Brandon-Luke L. Seagle; Thomas W. P. Friedl; Brigitte Rack; Krisztian Lato; Visnja Fink; Massimo Cristofanilli; Eric D. Donnelly; Wolfgang Janni; Shohreh Shahabi; et al. Association of Circulating Tumor Cell Status With Benefit of Radiotherapy and Survival in Early-Stage Breast Cancer. JAMA Oncology 2018, 4, e180163, 10.1001/jamaoncol.2018.0163.
- Anna-Maria Larsson; Sara Jansson; Pär-Ola Bendahl; Charlotte Levin Tykjaer Jörgensen; Niklas Loman; Cecilia Graffman; Lotta Lundgren; Kristina E. Aaltonen; L. Rydén; Longitudinal enumeration and cluster evaluation of circulating tumor cells improve prognostication for patients with newly diagnosed metastatic breast cancer in a prospective observational trial. Breast Cancer Research 2018, 20, 48, 10.1186/s13058-018-0976-0.
- Xiaoqiu Wu; Fangfei Li; Lei Dang; Chao Liang; Aiping Lu; Ge Zhang; RANKL/RANK System-Based Mechanism for Breast Cancer Bone Metastasis and Related Therapeutic Strategies. Frontiers in Cell and Developmental Biology 2020, 8, 76, 10.3389/fcell.2020.00076.
- Francesco Pantano; Elisabetta Rossi; Michele Iuliani; Antonella Facchinetti; Sonia Simonetti; Giulia Ribelli; Alice Zoccoli; Bruno Vincenzi; Giuseppe Tonini; Rita Zamarchi; et al. Dynamic changes of Receptor activator of nuclear factor-κB expression in Circulating Tumor Cells during Denosumab predict treatment effectiveness in Metastatic Breast Cancer. Scientific Reports 2020, 10, 1-12, 10.1038/s41598-020-58339-2.
- Jaeger, B.A.; Neugebauer, J.; Andergassen, U.; Melcher, C.; Schochter, F.; Mouarrawy, D.; Ziemendorff, G.; Clemens, M.; Abel, E.V.; Heinrich, G.; et al. The HER2 phenotype of circulating tumor cells in HER2-positive early breast cancer: A translational research project of a prospective randomized phase III trial. PLoS ONE 2017, 12, e0173593.
- Schochter, F.; Friedl, T.W.P.; deGregorio, A.; Krause, S.; Huober, J.; Rack, B.; Janni, W. Are Circulating Tumor Cells (CTCs) Ready for Clinical Use in Breast Cancer? An Overview of Completed and Ongoing Trials Using CTCs for Clinical Treatment Decisions. Cells 2019, 8, 1412.
- Kodahl, A.R.; Ehmsen, S.; Pallisgaard, N.; Jylling, A.M.B.; Jensen, J.D.; Laenkholm, A.V.; Knoop, A.S.; Ditzel, H.J. Correlation between circulating cell-free PIK3CA tumor DNA levels and treatment response in patients with PIK3CA-mutated metastatic breast cancer. Mol. Oncol. 2018, 12, 925–935.
- Beaver, J.A.; Jelovac, D.; Balukrishna, S.; Cochran, R.; Croessmann, S.; Zabransky, D.J.; Wong, H.Y.; Toro, P.V.; Cidado, J.; Blair, B.G.; et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin. Cancer Res. 2014, 20, 2643–2650.
- Chandarlapaty, S.; Chen, D.; He, W.; Sung, P.; Samoila, A.; You, D.; Bhatt, T.; Patel, P.; Voi, M.; Gnant, M.; et al. Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol. 2016, 2, 1310–1315.
- R. Charles Coombes; Karen Page; Raheleh Salari; Robert K. Hastings; Anne C. Armstrong; Samreen Ahmed; Simak Ali; Susan Jane Cleator; Laura M. Kenny; Justin Stebbing; et al. Personalized Detection of Circulating Tumor DNA Antedates Breast Cancer Metastatic Recurrence. Clinical Cancer Research 2019, 25, 4255-4263, 10.1158/1078-0432.ccr-18-3663.
- Bradon R. McDonald; Tania Contente-Cuomo; Stephen-John Sammut; Ahuva Odenheimer-Bergman; Brenda Ernst; Nieves Perdigones; Suet-Feung Chin; Maria Farooq; Rosa Mejia; Patricia A. Cronin; et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Science Translational Medicine 2019, 11, eaax7392, 10.1126/scitranslmed.aax7392.
- Miguel R Ossandon; Lokesh Agrawal; Eric J Bernhard; Barbara A Conley; Sumana M Dey; Rao L Divi; Ping Guan; Tracy G Lively; Tawnya C McKee; Brian S Sorg; et al. Circulating Tumor DNA Assays in Clinical Cancer Research. JNCI Journal of the National Cancer Institute 2018, 110, 929-934, 10.1093/jnci/djy105.
- Chetan Bettegowda; Mark Sausen; Rebecca J. Leary; Isaac Kinde; Yuxuan Wang; Nishant Agrawal; Bjarne R. Bartlett; Hao Wang; Brandon Luber; Rhoda M. Alani; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Science Translational Medicine 2014, 6, 224ra24-224ra24, 10.1126/scitranslmed.3007094.
- Heidi Schwarzenbach; Jan Stoehlmacher; Klaus Pantel; Eray Goekkurt; Detection and Monitoring of Cell-Free DNA in Blood of Patients with Colorectal Cancer. Annals of the New York Academy of Sciences 2008, 1137, 190-196, 10.1196/annals.1448.025.
- Devonshire, A.S.; Whale, A.S.; Gutteridge, A.; Jones, G.; Cowen, S.; Foy, C.A.; Huggett, J.F. Towards standardisation of cell-free DNA measurement in plasma: Controls for extraction efficiency, fragment size bias and quantification. Anal. Bioanal. Chem. 2014, 406, 6499–6512.
- Tangvarasittichai, O.; Jaiwang, W.; Tangvarasittichai, S. The plasma DNA concentration as a potential breast cancer screening marker. Indian J. Clin. Biochem. 2015, 30, 55–58.
- Wang, R.; Li, X.; Zhang, H.; Wang, K.; He, J. Cell-free circulating tumor DNA analysis for breast cancer and its clinical utilization as a biomarker. Oncotarget 2017, 8, 75742–75755.
- Kruger, D.T.; Jansen, M.; Konings, I.; Dercksen, W.M.; Jager, A.; Oulad Hadj, J.; Sleijfer, S.; Martens, J.W.M.; Boven, E. High ctDNA molecule numbers relate with poor outcome in advanced ER+, HER2- postmenopausal breast cancer patients treated with everolimus and exemestane. Mol. Oncol. 2020, 14, 490–503.
- Elazezy, M.; Joosse, S.A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput. Struct. Biotechnol. J. 2018, 16, 370–378.
- Freidin, M.B.; Freydina, D.V.; Leung, M.; Montero Fernandez, A.; Nicholson, A.G.; Lim, E. Circulating tumor DNA outperforms circulating tumor cells for KRAS mutation detection in thoracic malignancies. Clin. Chem. 2015, 61, 1299–1304.
- Vymetalkova, V.; Cervena, K.; Bartu, L.; Vodicka, P. Circulating Cell-Free DNA and Colorectal Cancer: A Systematic Review. Int. J. Mol. Sci. 2018, 19, 3356.
- Jenkins, S.; Yang, J.C.; Ramalingam, S.S.; Yu, K.; Patel, S.; Weston, S.; Hodge, R.; Cantarini, M.; Janne, P.A.; Mitsudomi, T.; et al. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non-Small Cell Lung Cancer. J. Thorac. Oncol. 2017, 12, 1061–1070.
- Wu, Y.L.; Lee, V.; Liam, C.K.; Lu, S.; Park, K.; Srimuninnimit, V.; Wang, J.; Zhou, C.; Appius, A.; Button, P.; et al. Clinical utility of a blood-based EGFR mutation test in patients receiving first-line erlotinib therapy in the ENSURE, FASTACT-2, and ASPIRATION studies. Lung Cancer 2018, 126, 1–8.
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88.
- Clare Fiala; Eleftherios P. Diamandis; Utility of circulating tumor DNA in cancer diagnostics with emphasis on early detection. BMC Medicine 2018, 16, 1-10, 10.1186/s12916-018-1157-9.
- Michaela J. Higgins; Danijela Jelovac; Evan Barnathan; Brian Blair; Shannon Slater; Penny Powers; Jane Zorzi; Stacie C. Jeter; George R. Oliver; John Fetting; et al. Detection of Tumor PIK3CA Status in Metastatic Breast Cancer Using Peripheral Blood. Clinical Cancer Research 2012, 18, 3462-3469, 10.1158/1078-0432.ccr-11-2696.
- Martinez-Saez, O.; Chic, N.; Pascual, T.; Adamo, B.; Vidal, M.; Gonzalez-Farre, B.; Sanfeliu, E.; Schettini, F.; Conte, B.; Braso-Maristany, F.; et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res. 2020, 22, 45.
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930.
- Board, R.E.; Wardley, A.M.; Dixon, J.M.; Armstrong, A.C.; Howell, S.; Renshaw, L.; Donald, E.; Greystoke, A.; Ranson, M.; Hughes, A.; et al. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res. Treat. 2010, 120, 461–467.
- Dawson, S.J.; Tsui, D.W.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 368, 1199–1209.
- Garcia-Saenz, J.A.; Ayllon, P.; Laig, M.; Acosta-Eyzaguirre, D.; Garcia-Esquinas, M.; Montes, M.; Sanz, J.; Barquin, M.; Moreno, F.; Garcia-Barberan, V.; et al. Tumor burden monitoring using cell-free tumor DNA could be limited by tumor heterogeneity in advanced breast cancer and should be evaluated together with radiographic imaging. BMC Cancer 2017, 17, 210.
- Andre, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940.
- Reinert, T.; Saad, E.D.; Barrios, C.H.; Bines, J. Clinical Implications of ESR1 Mutations in Hormone Receptor-Positive Advanced Breast Cancer. Front. Oncol. 2017, 7, 26.
- Toy, W.; Shen, Y.; Won, H.; Green, B.; Sakr, R.A.; Will, M.; Li, Z.; Gala, K.; Fanning, S.; King, T.A.; et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. 2013, 45, 1439–1445.
- Schiavon, G.; Hrebien, S.; Garcia-Murillas, I.; Cutts, R.J.; Pearson, A.; Tarazona, N.; Fenwick, K.; Kozarewa, I.; Lopez-Knowles, E.; Ribas, R.; et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci. Transl. Med. 2015, 7, 313ra182.
- Allouchery, V.; Beaussire, L.; Perdrix, A.; Sefrioui, D.; Augusto, L.; Guillemet, C.; Sarafan-Vasseur, N.; Di Fiore, F.; Clatot, F. Circulating ESR1 mutations at the end of aromatase inhibitor adjuvant treatment and after relapse in breast cancer patients. Breast Cancer Res. 2018, 20, 40.
- O’Leary, B.; Hrebien, S.; Beaney, M.; Fribbens, C.; Garcia-Murillas, I.; Jiang, J.; Li, Y.; Huang Bartlett, C.; Andre, F.; Loibl, S.; et al. Comparison of BEAMing and Droplet Digital PCR for Circulating Tumor DNA Analysis. Clin. Chem. 2019, 65, 1405–1413.
- Oxnard, G.R.; Thress, K.S.; Alden, R.S.; Lawrance, R.; Paweletz, C.P.; Cantarini, M.; Yang, J.C.; Barrett, J.C.; Janne, P.A. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2016, 34, 3375–3382.
- Spoerke, J.M.; Gendreau, S.; Walter, K.; Qiu, J.; Wilson, T.R.; Savage, H.; Aimi, J.; Derynck, M.K.; Chen, M.; Chan, I.T.; et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat. Commun. 2016, 7, 11579.
- Couraud, S.; Vaca-Paniagua, F.; Villar, S.; Oliver, J.; Schuster, T.; Blanche, H.; Girard, N.; Tredaniel, J.; Guilleminault, L.; Gervais, R.; et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: A proof-of-concept study from BioCAST/IFCT-1002. Clin. Cancer Res. 2014, 20, 4613–4624.
- Narayan, A.; Carriero, N.J.; Gettinger, S.N.; Kluytenaar, J.; Kozak, K.R.; Yock, T.I.; Muscato, N.E.; Ugarelli, P.; Decker, R.H.; Patel, A.A. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 2012, 72, 3492–3498.
- Thomas Reinert; Tenna Vesterman Henriksen; Emil Christensen; Shruti Sharma; Raheleh Salari; Himanshu Sethi; Michael Knudsen; Iver Nordentoft; Hsin-Ta Wu; Antony S. Tin; et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncology 2019, 5, 1124-1131, 10.1001/jamaoncol.2019.0528.
- Christopher Abbosh; The TRACERx consortium; Nicolai J. Birkbak; Gareth A. Wilson; Mariam Jamal-Hanjani; Tudor Constantin; Raheleh Salari; John Le Quesne; David A. Moore; Selvaraju Veeriah; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446-451, 10.1038/nature22364.
- Emil Christensen; Karin Birkenkamp-Demtröder; Himanshu Sethi; Svetlana Shchegrova; Raheleh Salari; Iver Nordentoft; Hsin-Ta Wu; Michael Knudsen; Philippe Lamy; Sia Viborg Lindskrog; et al. Early Detection of Metastatic Relapse and Monitoring of Therapeutic Efficacy by Ultra-Deep Sequencing of Plasma Cell-Free DNA in Patients With Urothelial Bladder Carcinoma. Journal of Clinical Oncology 2019, 37, 1547-1557, 10.1200/jco.18.02052.
- L. De Mattos-Arruda; B. Weigelt; J. Cortes; H. H. Won; C. K. Y. Ng; P. Nuciforo; F.-C. Bidard; C. Aura; V. Peg; S. Piscuoglio; et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Annals of Oncology 2014, 25, 1729-1735, 10.1093/annonc/mdu239.
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367.
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell Mol. Life Sci. 2018, 75, 193–208.
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal. Transduct. Target. Ther. 2020, 5, 145.
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013, 32, 623–642.
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, 421.
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009, 284, 34211–34222.
- McAndrews, K.M.; Kalluri, R. Mechanisms associated with biogenesis of exosomes in cancer. Mol. Cancer 2019, 18, 52.
- Valerie S. LeBleu; Raghu Kalluri; Exosomes as a Multicomponent Biomarker Platform in Cancer. Trends in Cancer 2020, 6, 767-774, 10.1016/j.trecan.2020.03.007.
- Zhou, B.; Xu, K.; Zheng, X.; Chen, T.; Wang, J.; Song, Y.; Shao, Y.; Zheng, S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal. Transduct. Target. Ther. 2020, 5, 144.
- Logozzi, M.; Mizzoni, D.; Di Raimo, R.; Fais, S. Exosomes: A Source for New and Old Biomarkers in Cancer. Cancers (Basel) 2020, 12, 2566.
- Xiaoxue Zhou; Feng Xie; Lin Wang; Long Zhang; Suping Zhang; Meiyu Fang; Fangfang Zhou; The function and clinical application of extracellular vesicles in innate immune regulation. Cellular & Molecular Immunology 2020, 17, 323-334, 10.1038/s41423-020-0391-1.
- Jia, Y.; Chen, Y.; Wang, Q.; Jayasinghe, U.; Luo, X.; Wei, Q.; Wang, J.; Xiong, H.; Chen, C.; Xu, B.; et al. Exosome: Emerging biomarker in breast cancer. Oncotarget 2017, 8, 41717–41733.
- Meng, Y.; Sun, J.; Wang, X.; Hu, T.; Ma, Y.; Kong, C.; Piao, H.; Yu, T.; Zhang, G. Exosomes: A Promising Avenue for the Diagnosis of Breast Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033818821421.
- Hadi Valadi; Karin Ekström; Apostolos Bossios; Margareta Sjöstrand; James J Lee; Jan O L Ouml Tvall; Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature 2007, 9, 654-659, 10.1038/ncb1596.
- Bethany Hannafon; Yvonne D. Trigoso; Cameron L. Calloway; Yan D. Zhao; David H. Lum; Alana L. Welm; Zhizhuang Joe Zhao; Kenneth E. Blick; William C Dooley; Wei-Qun Ding; et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Research 2016, 18, 1-14, 10.1186/s13058-016-0753-x.
- Corinna Eichelser; Isabel Stückrath; Volkmar Müller; Karin Milde-Langosch; Harriet Wikman; Klaus Pantel; Heidi Schwarzenbach; Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 2014, 5, 9650-9663, 10.18632/oncotarget.2520.
- Ines Stevic; Volkmar Müller; Karsten Weber; Peter A. Fasching; Thomas Karn; Frederic Marmé; Christian Schem; Elmar Stickeler; Carsten Denkert; Marion Van Mackelenbergh; et al. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Medicine 2018, 16, 1-16, 10.1186/s12916-018-1163-y.
- Alba Rodríguez-Martínez; Diego De Miguel-Pérez; Francisco Gabriel Ortega; José Luis García-Puche; Inmaculada Robles-Fernández; José Expósito; Jordi Martorell-Marugán; Pedro Carmona-Sáez; María Carmen Garrido-Navas; Christian Rolfo; et al. Exosomal miRNA profile as complementary tool in the diagnostic and prediction of treatment response in localized breast cancer under neoadjuvant chemotherapy. Breast Cancer Research 2019, 21, 1-9, 10.1186/s13058-019-1109-0.
- Aiko Sueta; Yutaka Yamamoto; Mai Tomiguchi; Takashi Takeshita; Mutsuko Yamamoto-Ibusuki; Hirotaka Iwase; Differential expression of exosomal miRNAs between breast cancer patients with and without recurrence. Oncotarget 2017, 8, 69934-69944, 10.18632/oncotarget.19482.
- Mio Yoshikawa; Hisae Iinuma; Yasuko Umemoto; Takako Yanagisawa; Akiko Matsumoto; Hiromitsu Jinno; Exosome‑encapsulated microRNA‑223‑3p as a minimally invasive biomarker for the early detection of invasive breast cancer. Oncology Letters 2018, 15, 9584-9592, 10.3892/ol.2018.8457.
- Marc Hirschfeld; Gerta Rücker; Daniela Weiß; Kai Berner; Andrea Ritter; Markus Jäger; Thalia Erbes; Urinary Exosomal MicroRNAs as Potential Non-invasive Biomarkers in Breast Cancer Detection. Molecular Diagnosis & Therapy 2020, 24, 215-232, 10.1007/s40291-020-00453-y.
- Marta Rodríguez; Javier Silva; Alberto Herrera; Mercedes Herrera; Cristina Peña; Paloma Martín; Beatriz Gil-Calderón; María Jesús Larriba; Mª José Coronado; Beatriz Soldevilla; et al. Exosomes enriched in stemness/metastatic-related mRNAS promote oncogenic potential in breast cancer. Oncotarget 2015, 6, 40575-40587, 10.18632/oncotarget.5818.
- Su-Jin Yang; Dan-Dan Wang; Jian Li; Han-Zi Xu; Hong-Yu Shen; Xiu Chen; Si-Ying Zhou; Shan-Liang Zhong; Jian-Hua Zhao; Jinhai Tang; et al. Predictive role of GSTP1-containing exosomes in chemotherapy-resistant breast cancer. Gene 2017, 623, 5-14, 10.1016/j.gene.2017.04.031.
- Marzia Del Re; Ilaria Bertolini; Stefania Crucitta; Lorenzo Fontanelli; Eleonora Rofi; Claudia De Angelis; Lucrezia Diodati; Diletta Cavallero; Giulia Gianfilippo; Barbara Salvadori; et al. Overexpression of TK1 and CDK9 in plasma-derived exosomes is associated with clinical resistance to CDK4/6 inhibitors in metastatic breast cancer patients. Breast Cancer Research and Treatment 2019, 178, 57-62, 10.1007/s10549-019-05365-y.
- Shicong Tang; Kai Zheng; Yiyin Tang; Zhen Li; Tianning Zou; Dequan Liu; Overexpression of serum exosomal HOTAIR is correlated with poor survival and poor response to chemotherapy in breast cancer patients. Journal of Biosciences 2019, 44, 37, 10.1007/s12038-019-9861-y.
- Guobin Zhong; Keqiong Wang; Jiawei Li; Shuzhe Xiao; Wei Wei; Jianlun Liu; Determination of Serum Exosomal H19 as a Noninvasive Biomarker for Breast Cancer Diagnosis. OncoTargets and Therapy 2020, 13, 2563-2571, 10.2147/ott.s243601.
- Huaying Dong; Wei Wang; Ru Chen; Yu Zhang; Kejian Zou; Mulin Ye; Xionghui He; Fan Zhang; Jing Han; Exosome-mediated transfer of lncRNA‑SNHG14 promotes trastuzumab chemoresistance in breast cancer. International Journal of Oncology 2018, 53, 1013-1026, 10.3892/ijo.2018.4467.
- Xiaodan Wang; Weiliang Zhong; Jingya Bu; Yuzhong Li; Ruihua Li; Rong Nie; Chenyang Xiao; Keli Ma; Xiaohua Huang; Ying Li; et al. Exosomal protein CD82 as a diagnostic biomarker for precision medicine for breast cancer. Molecular Carcinogenesis 2019, 58, 674-685, 10.1002/mc.22960.
- Aled Clayton; Attilla Turkes; Hossein Navabi; Malcolm D. Mason; Zsuzsanna Tabi; Induction of heat shock proteins in B-cell exosomes. Journal of Cell Science 2005, 118, 3631-3638, 10.1242/jcs.02494.
- Daniel R. Ciocca; Stuart K. Calderwood; Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress and Chaperones 2005, 10, 86-103, 10.1379/csc-99r.1.
- Gaëtan Chanteloup; Marine Cordonnier; Nicolas Isambert; Aurélie Bertaut; Alice Hervieu; Audrey Hennequin; Maxime Luu; Sylvie Zanetta; Bruno Coudert; Leila Bengrine; et al. Monitoring HSP70 exosomes in cancer patients’ follow up: a clinical prospective pilot study. Journal of Extracellular Vesicles 2020, 9, 1766192, 10.1080/20013078.2020.1766192.
- Moon, P.G.; Lee, J.E.; Cho, Y.E.; Lee, S.J.; Chae, Y.S.; Jung, J.H.; Kim, I.S.; Park, H.Y.; Baek, M.C. Fibronectin on circulating extracellular vesicles as a liquid biopsy to detect breast cancer. Oncotarget 2016, 7, 40189–40199.
- Moon, P.G.; Lee, J.E.; Cho, Y.E.; Lee, S.J.; Jung, J.H.; Chae, Y.S.; Bae, H.I.; Kim, Y.B.; Kim, I.S.; Park, H.Y.; et al. Identification of Developmental Endothelial Locus-1 on Circulating Extracellular Vesicles as a Novel Biomarker for Early Breast Cancer Detection. Clin. Cancer Res. 2016, 22, 1757–1766.
- Lee, J.E.; Moon, P.G.; Cho, Y.E.; Kim, Y.B.; Kim, I.S.; Park, H.; Baek, M.C. Identification of EDIL3 on extracellular vesicles involved in breast cancer cell invasion. J. Proteom. 2016, 131, 17–28.
- Khan, S.; Bennit, H.F.; Turay, D.; Perez, M.; Mirshahidi, S.; Yuan, Y.; Wall, N.R. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer 2014, 14, 176.
- Sonia A. Melo; Linda B. Luecke; Christoph Kahlert; Agustín Fernández Fernández; Seth T. Gammon; Judith Kaye; Valerie S. LeBleu; Elizabeth A. Mittendorf; Juergen Weitz; Nuh N Rahbari; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177-182, 10.1038/nature14581.
- Rupp, A.K.; Rupp, C.; Keller, S.; Brase, J.C.; Ehehalt, R.; Fogel, M.; Moldenhauer, G.; Marme, F.; Sultmann, H.; Altevogt, P. Loss of EpCAM expression in breast cancer derived serum exosomes: Role of proteolytic cleavage. Gynecol. Oncol. 2011, 122, 437–446.
- Fang, S.; Tian, H.; Li, X.; Jin, D.; Li, X.; Kong, J.; Yang, C.; Yang, X.; Lu, Y.; Luo, Y.; et al. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PLoS ONE 2017, 12, e0175050.
- I-Hsuan Chen; Liang Xue; Chuan-Chih Hsu; Juan Sebastian Paez Paez; Li Pan; Hillary Andaluz; Michael K. Wendt; Anton B. Iliuk; Jian‐Kang Zhu; W. Andy Tao; et al. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proceedings of the National Academy of Sciences 2017, 114, 3175-3180, 10.1073/pnas.1618088114.
- Maria Galli De Amorim; Gustavo Ribeiro Fernandes; Paulo Oliveira; Daniel Martins-De-Souza; Emmanuel Dias-Neto; Diana Noronha Nunes; The overexpression of a single oncogene (ERBB2/HER2) alters the proteomic landscape of extracellular vesicles. PROTEOMICS 2014, 14, 1472-1479, 10.1002/pmic.201300485.
- Stamatia Rontogianni; Eleni Synadaki; Bohui Li; Marte C. Liefaard; Esther H. Lips; Jelle Wesseling; Wei Wu; Maarten Altelaar; Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Communications Biology 2019, 2, 1-13, 10.1038/s42003-019-0570-8.
- Valentina Ciravolo; Veronica Huber; Gaia C. Ghedini; Elisabetta Venturelli; Francesca Bianchi; M. Campiglio; Daniele Morelli; Antonello Villa; Pamela Della Mina; Sylvie Menard; et al. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. Journal of Cellular Physiology 2011, 227, 658-667, 10.1002/jcp.22773.
- Ning, K.; Wang, T.; Sun, X.; Zhang, P.; Chen, Y.; Jin, J.; Hua, D. UCH-L1-containing exosomes mediate chemotherapeutic resistance transfer in breast cancer. J. Surg. Oncol. 2017, 115, 932–940.
- Ma, X.; Chen, Z.; Hua, D.; He, D.; Wang, L.; Zhang, P.; Wang, J.; Cai, Y.; Gao, C.; Zhang, X.; et al. Essential role for TrpC5-containing extracellular vesicles in breast cancer with chemotherapeutic resistance. Proc. Natl. Acad. Sci. USA 2014, 111, 6389–6394.
- Wang, T.; Ning, K.; Lu, T.X.; Sun, X.; Jin, L.; Qi, X.; Jin, J.; Hua, D. Increasing circulating exosomes-carrying TRPC5 predicts chemoresistance in metastatic breast cancer patients. Cancer Sci. 2017, 108, 448–454.
- Bossuyt, P.M. Clinical validity: Defining biomarker performance. Scand. J. Clin. Lab. Invest. Suppl. 2010, 242, 46–52.
- Pletcher, M.J.; Pignone, M. Evaluating the clinical utility of a biomarker: A review of methods for estimating health impact. Circulation 2011, 123, 1116–1124.
- Isabel Heidrich; Lucija Ačkar; Parinaz Mossahebi Mohammadi; Klaus Pantel; Liquid biopsies: Potential and challenges. International Journal of Cancer 2020, 148, 528-545, 10.1002/ijc.33217.
- Kelly Rae Chi; The tumour trail left in blood. Nature 2016, 532, 269-271, 10.1038/532269a.
- Guislaine Barrière; Pietro Fici; Giulia Gallerani; Francesco Fabbri; Wainer Zoli; Michel Rigaud; Circulating tumor cells and epithelial, mesenchymal and stemness markers: characterization of cell subpopulations. Annals of Translational Medicine 2014, 2, 109, 10.3978/j.issn.2305-5839.2014.10.04.
- Prieto-Garcia, E.; Diaz-Garcia, C.V.; Garcia-Ruiz, I.; Agullo-Ortuno, M.T. Epithelial-to-mesenchymal transition in tumor progression. Med. Oncol. 2017, 34, 122.
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. Emt: 2016. Cell 2016, 166, 21–45.
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629.
- Hyun, K.A.; Koo, G.B.; Han, H.; Sohn, J.; Choi, W.; Kim, S.I.; Jung, H.I.; Kim, Y.S. Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget 2016, 7, 24677–24687.
- Michail Ignatiadis; Françoise Rothé; Carole Chaboteaux; Virginie Durbecq; Ghizlane Rouas; Carmen Criscitiello; Jessica Metallo; Naima Kheddoumi; Sandeep K. Singhal; Stefan Michiels; et al. HER2-Positive Circulating Tumor Cells in Breast Cancer. PLOS ONE 2011, 6, e15624, 10.1371/journal.pone.0015624.
- Min-Sun Cho; Chul Hwan Park; Sungsoo Lee; Heae Surng Park; Clinicopathological parameters for circulating tumor DNA shedding in surgically resected non-small cell lung cancer with EGFR or KRAS mutation. PLOS ONE 2020, 15, e0230622, 10.1371/journal.pone.0230622.
- Gustav Johansson; Daniel Andersson; Stefan Filges; Junrui Li; A. Muth; Tony E. Godfrey; Anders Ståhlberg; Considerations and quality controls when analyzing cell-free tumor DNA. Biomolecular Detection and Quantification 2019, 17, 100078, 10.1016/j.bdq.2018.12.003.
- Daniel Stetson; Ambar Ahmed; Xing Xu; Barrett R.B. Nuttall; Tristan J. Lubinski; Justin H. Johnson; J. Carl Barrett; Brian A. Dougherty; Orthogonal Comparison of Four Plasma NGS Tests With Tumor Suggests Technical Factors are a Major Source of Assay Discordance. JCO Precision Oncology 2019, null, 1-9, 10.1200/po.18.00191.
- Srujan Gandham; Xianyi Su; Jacqueline Wood; Angela L. Nocera; Sarath Chandra Alli; Lara Milane; Alan Zimmerman; Mansoor Amiji; Alexander R. Ivanov; Technologies and Standardization in Research on Extracellular Vesicles. Trends in Biotechnology 2020, 38, 1066-1098, 10.1016/j.tibtech.2020.05.012.
- Marta Tellez-Gabriel; Minique Heymann; Exosomal lncRNAs: the newest promising liquid biopsy. Cancer Drug Resistance 2019, null, null, 10.20517/cdr.2019.69.
- Zhong Ye; Chun Wang; Shaogui Wan; Zhaomei Mu; Zhenchao Zhang; Maysa M. Abu-Khalaf; Frederick M. Fellin; Daniel P. Silver; Manish Neupane; Rebecca J. Jaslow; et al. Association of clinical outcomes in metastatic breast cancer patients with circulating tumour cell and circulating cell-free DNA. European Journal of Cancer 2019, 106, 133-143, 10.1016/j.ejca.2018.10.012.
