The human brainstem plays important roles in maintaining basic vital functions. In comparison with brain functional magnetic resonance imaging (fMRI), only a few fMRI studies investigating the brainstem have been reported because of a number of technical challenges. This entry briefly introduces technical difficulties, recent advances, and further directions of brainstem fMRI in humans.
- brainstem
- fMRI
- human
1. Introduction
The human brainstem plays important roles in maintaining basic vital functions, e.g., respiration, swallowing, and walking. The brainstem nuclei play essential roles in the regulation of behavior and cognition via cortical/subcortical projections and nerves originating from the nuclei [1]. For instance, the brainstem nuclei for vagal motor innervation are the nucleus ambiguus and dorsal motor nucleus of the vagus, and the final motoneurons are the ganglia in the lower respiratory tract and lung, the cardiac ganglion, and enteric neurons in the abdominal organs. The vagus nerve supplies the pharynx, larynx, and esophagus (general motor and sensory); the thorax and abdomen (visceral); and the thoracic and abdominal organs (parasympathetic nerve endings). Parasympathetic neurons in the dorsal vagus motor nucleus innervate the ganglia in the gastrointestinal wall and other abdominal organs. The motor nucleus modulates visceral motor function, e.g., activities of the gastrointestinal smooth muscle. The brainstem nucleus for the vagal sensory innervation is the nucleus tractus solitarius. As for the swallowing function, most of the motor or sensory nerves supplying to the pharynx originate from the pharyngeal plexus. The plexus consists of the branches of the glossopharyngeal nerve, vagus nerves, and superior cervical sympathetic ganglia. The pharyngeal muscles are innervated from the pharyngeal plexus (through the vagal pharyngeal branch), except the stylopharyngeus (innervated by the glossopharyngeal nerve) [2][3].
However, unlike a large amount of brain functional magnetic resonance imaging (fMRI) research, only a much less number of fMRI studies investigating the brainstem have been reported until now due to various technical challenges. This entry briefly introduces technical difficulties, recent advances, and further directions of brainstem fMRI in humans.
2. Technical Challenges of Brainstem fMRI
2.1. Motion Artifacts
Motion artifacts of the brainstem are a big challenge for fMRI research. Movements of brain tissues during fMRI scanning lead to displacements of voxels and thus influence voxel intensity. Motion artifacts stem from a number of sources including head movements, movements of the brainstem itself, cerebrospinal fluid (CSF) flux, and blood flow [4].Head movements during MRI scanning cannot be totally avoided and are often treated with a rigid-body motion correction approach. In comparison with the voxel size, even a small amplitude of head motion can lead to the variation of voxel intensity in blood-oxygen-level-dependent (BOLD) fMRI images, thereby interfering with the detection of signal changes originating from cerebral activities. Motion correction tools including those in Automated Image Registration (AIR), Analysis of Functional NeuroImages (AFNI), FMRIB Software Library (FSL), and Statistical Parametric Mapping (SPM) can lead to improvements in the magnitude (up to 20%) and cluster size (up to 100%) of brain activations. Most motion correction methods apply a rigid-body algorithm, assuming that the shape of the brain is constant across volumes collected at different time points for the same subject. This rigid-body approach corrects translational and rotational motion [5].Head motion correction may lead to acceptable results for motions of cerebral hemispheres and cerebellum if the amplitude of head movements fluctuates slightly. However, similar to the spinal cord, human brainstem has a thin profile. It is surrounded by cerebrospinal fluid, which cannot restrain small extents of brainstem motion relative to other parts of the brain, or motion of one part relative to other parts of the brainstem. The brainstem is therefore easily distorted by movements of various sources, in addition to rigid-body translational and rotational motions. The caudal part of the brainstem connects the cervical spinal cord, a structure nearly “floating” in the vertebral canal and highly influenced by motion [6]; movements of the cervical spinal cord result in brainstem motions, such as the elongation-contraction movement, which enhances distortion of the brainstem [7].Both cardiac and pulmonary activities lead to brainstem distortions. Respiration evokes brainstem oscillation, in which the cranial and caudal parts of brainstem displacements do not synchronize but correspond with different respiratory phases (expiration or inspiration) [8][9][10]. Ventilatory chest movement may drive brainstem movements through spinal cord movements (e.g., chest motion→vertebral canal and nerve roots from the spinal cord→spinal cord→brainstem) or through CSF motion driven by respiratory circles.Brainstem motion originated from cardiac activities also presents non synchronized displacements between caudal and rostral structures of the brainstem [11]. The cardiac activity leads to pulsatility of arteries supplying the brainstem including the basilar artery and vertebral artery, which causes brainstem movements.Note that distortion includes changes in both shape and size of the brainstem, which cannot be corrected by using a rigid-body model.
2.2. Physiological Noise
Here, the physiological noise is introduced as factors except brainstem motion, although the physiological noise and brainstem motion may stem from the same sources (e.g., cardiac and pulmonary activities). For brainstem fMRI, both cardiac and pulmonary activities are also main sources of physiologically-originated noise, that is, BOLD signal changes in acquired images caused by the subject’s physiological factors but not brain activities. For example, the respiratory cycle is associated with lung volume changes and chest wall movements, which leads to a varying main static magnetic field (B0) originating from variations of oxygen concentration/spatial distribution in the lungs. Such respiratory-induced instability of BOLD-fMRI signals in the brainstem and cervical spinal cord is higher than the instability in more cranial regions of the brain [4]. In addition, the respiratory cycle also has impact on arterial CO2 partial pressure. The cardiac cycle influences cerebral blood flow and volume [12]. BOLD signal changes originating from these factors are noise contaminating signal variations induced by brain function changes.
2.3. Susceptibility-Induced BOLD Signal Losses/Distortions
The magnetic susceptibility of an object indicates its tendency to be magnetized when placed into a magnetic field. At 37 °C, the susceptibility of water is −9.05 × 10−6, and the susceptibilities of most human tissues are within ±10–20% of this value. At 20 °C and normal pressure (1013 mbar), the susceptibility of air is 0.36 × 10−6 [13][14]. Susceptibility artifacts originate from the difference in the magnetic susceptibility arising at an air-tissue or metal implant-tissue interface, resulting in changes in both location and intensity of voxels.
In BOLD fMRI, susceptibility-related field inhomogeneities near the interface of tissues/air with strong different magnetic susceptibilities produce signal losses and/or image distortions. These susceptibility-induced BOLD signal losses and distortions influence sensitivity of spinal cord fMRI and brainstem fMRI [15]. Even in less-affected regions such as the primary motor hand area, voxel shift values in raw echo-planar imaging (EPI) images may reach 5 mm [16].
The sphenoid and the air-filled sphenoid sinus are adjacent to the brainstem, causing magnetic susceptibility-induced BOLD sensitivity losses of the medulla and especially ventral pons [17]. The FieldMap toolbox calculates an unwrapped field map with collected field maps during data acquisition, converts the created field map to the voxel displacement map, and then unwarps geometrically distorted BOLD-fMRI images. This method or similar tools are helpful but can hardly achieve a satisfactory correction for the ventral part of the pons (Figure 1) [18].
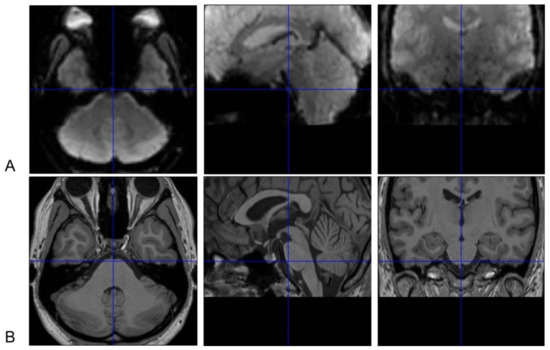
Figure 1. Susceptibility-induced BOLD signals loss of the ventral pons in functional magnetic resonance imaging (fMRI) images. Normalized brainstem fMRI (A) and T1 images (B) of one subject in [18] are presented. The ventral pons appears to be “dark” in fMRI images because of susceptibility-induced loss of BOLD signals after correction with field maps. Thus, BOLD signal changes in this area cannot be reliably measured. These images are displayed with the SPM12 software (https://www.fil.ion.ucl.ac.uk/spm). The crosshairs in BOLD and T1 images indicate the same point in the Montreal Neurological Institute (MNI) space. BOLD signal losses are also obvious in the medial side of the temporal lobe besides in the ventral pons.
2.4. Thermal Noise and Low-Frequency Drift
The temporal signal-to-noise ratio (tSNR) is a ratio between the average BOLD signal intensity during an fMRI session and the standard deviation of the BOLD signal intensity of the session. The brainstem presents much lower tSNR than cerebral and cerebellar tissues, demonstrating relatively small magnitudes of BOLD signal changes compared to noise in the brainstem [12].
Thermal fluctuations of scanned subjects and MR electronics during an fMRI session are sources of noise. In comparison with the cortex, a longer distance of the brainstem from the MR coil reduces coil sensitivity, which may increase impacts from the thermal noise at a higher spatial resolution [4]. SNR driven by thermal fluctuations varies linearly with voxel sizes [12]. A higher spatial resolution of fMRI images (i.e., a smaller voxel size) increases the capability to differentiate anatomical details but reduces the SNR. A large voxel size can increase SNR but reduce tSNR gains due to worse impacts from physiologic noise. To acquire the highest SNR at the cost of the minimum loss in spatial resolution, suggested voxel sizes for gray matter, white matter, and cerebral CSF at 3T MR images have been provided as 1.8 mm3, 2.1 mm3, and 1.4 mm3, respectively [19].
The baseline of BOLD signal drifts during an fMRI session, which is usually considered to originate from the instability of MR instruments [20]. The signal drift may be caused by intrinsic brain physiology [21] whereas the impact of this factor on the brainstem remains unknown.
2.5. Localizing Brainstem Nuclei
Prior to group data analysis, individual MRI data needs to be transformed to match a template (e.g., MNI standard brains) through affine registration, e.g., “normalization” with SPM software or linear intra- and inter-modal brain image registration with FMRIB’s Linear Image Registration Tool. Such a process aims to facilitate accurate coregistration of anatomical structures across all subjects. After normalization, the locations, scopes, and sizes of brain regions in each subject correspond to those in the template. The MNI or other templates can therefore establish a common three-dimensional coordinate system as a reference to consistently communicate and compare the functional anatomy across studies [22][23]. Since a number of nuclei are densely situated in the brainstem, small coregistration errors result in increased variance/decreased significance in detected activations [24].
The cross-sectional size of the brainstem is much smaller than that of brain hemispheres, and a number of brainstem nuclei have very small volumes [25]. Thus, differentiating brainstem nuclei is much harder than parcellating brain areas in cerebral hemispheres during fMRI data analysis. Cytoarchitectonic probability maps have been developed from the analysis of post-mortem human brain to correlate microscopically anatomical data and functional imaging data of the cerebral cortex [26]. However, there is no cytoarchitectonic probability map for human brainstem by now, and thus it is impossible to automatically determine anatomical correspondence to a brainstem template. Manually localizing/labeling brainstem nuclei is a common approach to establish anatomical correspondence in brainstem fMRI studies. Such a manual process may be time and resource consuming and lack a consistent standard.
3. Advances in Brainstem fMRI
After motion correction, brainstem and spinal cord fMRI data still need to remove effects of physiological noise by using physiological (e.g., pulse and respiratory) recordings as regressors in the General Linear Model. This approach needs to take physiological recordings during fMRI scanning (e.g., using a pulse oximeter and a respiratory bellows [27]), better with the concurrent recording of scanner triggers [28].
Cardiac gating can be applied to reduce cardiogenic noise when collecting brainstem fMRI data. However, this technique has several disadvantages: (1) Fluctuations in the duration of the repetition time (TR) induced by heart rate variability cause temporal fluctuations of the intensity in fMRI images. (2) Due to non-constant TRs in cardiac gating, a mean TR is needed during data analysis, which may not be supported by commonly-used fMRI softwares. (3) The cardiac gating method leads to relatively longer TR values, e.g., ranged approximately between 3.6 s and 4.6 s (an average value of 4.1 s) [7], which limits the time resolution of fMRI. Thus, collecting data with a standard fMRI sequence has been advocated. Independent component analysis finds that physiological noise is mainly detected at the junction area between the brainstem tissues and the adjacent CSF and blood vessels [4]. Brainstem masks are therefore used to prevent impacts driven by noise from the interface between the brainstem and surrounding tissues. The masked analysis has been applied successfully to resting-state brainstem fMRI data [29] and then used to analyze task-induced activations in the brainstem [18]. These brainstem masks are manually delineated on the basis of mean BOLD images, whereas a CSF mask can be automatically generated to extract nuisance regressors for the aCompCor-based method. The aCompCor approach regresses signal changes in the CSF out of BOLD signals in the statistical model because these signal changes are unlikely elicited by neuronal activities [30].
To reduce effects of small coregistration errors on detecting brainstem activations, the 2-stage mask guided registration technique has been developed to improved brainstem coregistration, and proven to generate a more consistent overlap of individual brainstem borders when compared to whole-brain coregistration methods [24]. Smoothing with a 4.5 mm-kernel combined with this mask guided registration method is proven to be sensitive for detecting activation of brainstem nuclei [7]. When using FSL normalization, activation of the trigeminal motor nucleus was detected by using 3 or 4.5 mm-kernel smoothing but not 6 mm-kernel smoothing [7]. Evidently, smoothing with a large kernel may “blur” localized activations and thus lead to loss in sensitivity. On the other hand, no smoothing or using small kernels may not well enable inter-subject consistency, which also reduces sensitivity. For example, compared to a 3 or 4 mm-kernel, smoothing with a 5 mm-kernel resulted in larger task-evoked activations in the brainstem [18]. No smoothing also resulted in more dispersed activation clusters in cortical areas and therefore appeared to be less sensitive in comparison with smoothing with a 6 mm-kernel [31].
To achieve brainstem segmentation, tissue probability maps for four classes of brainstem tissues (three gray-matters and one white-matter) acquired with 3 T MRI scanners have been generated, based on a brain specimen of a 57-year-old male subject. These four classes of brainstem tissues are well consistent with brainstem anatomy shown in MR microscopy at 9.4 T [32]. This method can be used to quantitatively analyze the internal architecture of the brainstem [33]. To study functional connectivity between the brainstem and cortical regions [34] or between different nuclei of the brainstem [35][36], it is necessary to define the exact scopes of brainstem nuclei. The cuneiform nucleus (CN) is located at the lateral side of the periaqueductal gray matter, and the pedunculopontine nucleus (PPN) is located at the lateral side of the CN. The CN controls high-speed locomotion whereas the PPN mediates slow/exploratory locomotion in vertebrates [37][38]. Localizing individual nuclei, e.g., the CN or the PPN on the basis of a histochemically defined atlas [25] has also been reported [18]. With higher magnetic susceptibility, iron-rich structures including the red nucleus and substantia nigra can be defined by using quantitative susceptibility mapping [39]. Visual correspondence between detected brainstem activations and an atlas has also been used to estimate the location of brainstem nuclei, e.g., in a study to explore the effect of stimulating the auricular branch of the vagus nerve on the nucleus tractus solitarii in humans [40]. Ultrahigh field (7T) MR scanners can provide a high spatial resolution (e.g., 1.2 mm isotropic voxels) and strong tissue contrast in BOLD images, thereby reducing misalignments between functional and anatomical MR images [41].
4. Further Directions
High quality of fMRI data is of primary importance for a research. Head movements during scanning are the main issue for brain fMRI studies. The real-time amplitude of head motion can be monitored [42] as an index to restart fMRI scanning. Notably, such real-time monitoring and a routine motion correction procedure in the pipeline of fMRI data analysis target movements of the entire head. However, the acceptable amplitude of head motion during scanning, e.g., less than 1- or 2-mm translation in x, y, and z axes, does not necessarily correspond to the same amplitude of brainstem motion. An accurate estimation of the extent of brainstem motion should originate from calculating displacements of the brainstem rather than the entire brain. As a precondition of such estimation, precise automatic segmentation of the brainstem and its surrounding tissues may be needed, similar to a skull-stripping process for brain images.
For the cervical spinal cord, slice-wise motion correction is better than volume-wise correction since the distortion of spinal cord driven by the cardiac and respiratory cycle varies along the cranial-caudal direction [43]. Similarly, because of nonsynchronized motions between different parts of the brainstem [8][9][10] and more localized impacts such as artery pulsations, rigid-body alignments across volumes of fMRI images (e.g., intrasubject registration) may not achieve accurate motion correction. Linear/nonlinear algorithms (e.g., affine models) [44] are therefore worthy to be tried to correct deformation of the brainstem. In previous studies, improvements after modeling of physiological noise [43] or using masks to exclude brainstem margins [18][29] may also benefit from eliminating effects of residual brainstem distortions after motion-correction process.
Brain [26][45] and spinal cord atlases [46][47] provide standard reference spaces for projecting functional changes to anatomical structures in fMRI studies. Although the brainstem atlas has not been developed, the relationship of histological staining-structure MRI [48] or detailed histological staining of the brainstem [25] have been reported. A brainstem template is needed to achieve an accurate corresponding relationship between histologically defined nuclei/white matter fiber and high-resolution MRI. Notably, brain templates generated from different populations (Caucasian and Chinese people) exhibit dramatic differences in supramarginal gyri and inferior frontal gyri in terms of deformation variability, which can reduce the performance of the brain segmentation and registration when a mismatched template is applied during spatial normalization. The sample size during template constructing process also influences the deformation variability in some brain regions, i.e., the frontoparietal control network and dorsal attention network [49].
Subject-specific functional organization of brainstem nuclei should also be noted. Inter-subject variability in functional mapping of cortical regions has been identified. Such cortical functional regions present strong inter-subject variation in size, location, and connectivity. Furthermore, the size and location of the cortical regions as well as the resting-state functional connectivity among these regions are related to the performance of human behavior. Interestingly, the inter-subject differences in size, location, or connectivity of cortical regions can bring dissociated information to explain the behavior of subjects [50][51]. Therefore, it is worthy to determine individual-specific functional organization of brainstem nuclei and evaluate the relationship between inter-subject variation and behavioral performance.
The entry is from https://www.mdpi.com/2309-3366/1/1/3/htm
References
- Beissner, F.; Baudrexel, S. Investigating the human brainstem with structural and functional MRI. Front. Hum. Neurosci. 2014, 8, 116.
- Ramachandran, V.S. (Ed.) Encyclopedia of the Human Brain, 1st ed.; Academic Press: San Diego, CA, USA, 2002; Volume 4, pp. 543–906.
- Standring, S. Gray’s Anatomy; Churchill Livingstone: London, UK, 2004; pp. 441–724.
- Sclocco, R.; Beissner, F.; Bianciardi, M.; Polimeni, J.R.; Napadow, V. Challenges and opportunities for brainstem neuroimaging with ultrahigh field MRI. Neuroimage 2018, 168, 412–426.
- Oakes, T.R.; Johnstone, T.; Walsh, K.O.; Greischar, L.L.; Alexander, A.L.; Fox, A.S.; Davidson, R.J. Comparison of fMRI motion correction software tools. Neuroimage 2005, 28, 529–543.
- Wei, P.; Li, J.; Gao, F.; Ye, D.; Zhong, Q.; Liu, S. Resting state networks in human cervical spinal cord observed with fMRI. Eur. J. Appl. Physiol. 2010, 108, 265–271.
- Beissner, F.; Deichmann, R.; Baudrexel, S. fMRI of the brainstem using dual-echo EPI. Neuroimage 2011, 55, 1593–1599.
- Whedon, J.M.; Glassey, D. Cerebrospinal fluid stasis and its clinical significance. Altern. Ther. Health Med. 2009, 15, 54–60.
- Budgell, B.S.; Bolton, P.S. Cerebrospinal fluid pressure in the anesthetized rat. J. Manip. Physiol. Ther. 2007, 30, 351–356.
- Maier, S.E.; Hardy, C.J.; Jolesz, F.A. Brain and cerebrospinal fluid motion: Real-time quantification with M-mode MR imaging. Radiology 1994, 193, 477–483.
- Enzmann, D.R.; Pelc, N.J. Brain motion: Measurement with phase-contrast MR imaging. Radiology 1992, 185, 653–660.
- Brooks, J.C.; Faull, O.K.; Pattinson, K.T.; Jenkinson, M. Physiological noise in brainstem FMRI. Front. Hum. Neurosci. 2013, 7, 623.
- Petersch, B.; Bogner, J.; Fransson, A.; Lorang, T.; Pötter, R. Effects of geometric distortion in 0.2T MRI on radiotherapy treatment planning of prostate cancer. Radiother. Oncol. 2004, 71, 55–64.
- Schenck, J.F. The role of magnetic susceptibility in magnetic resonance imaging: MRI magnetic compatibility of the first and second kinds. Med. Phys. 1996, 23, 815–850.
- Cordes, D.; Turski, P.A.; Sorenson, J.A. Compensation of susceptibility-induced signal loss in echo-planar imaging for functional applications. Magn. Reson. Imaging 2000, 18, 1055–1068.
- Lima Cardoso, P.; Dymerska, B.; Bachratá, B.; Fischmeister, F.P.S.; Mahr, N.; Matt, E.; Trattnig, S.; Beisteiner, R.; Robinson, S.D. The clinical relevance of distortion correction in presurgical fMRI at 7T. Neuroimage 2018, 168, 490–498.
- Weiskopf, N.; Hutton, C.; Josephs, O.; Deichmann, R. Optimal EPI parameters for reduction of susceptibility-induced BOLD sensitivity losses: A whole-brain analysis at 3 T and 1.5 T. Neuroimage 2006, 33, 493–504.
- Wei, P.; Zou, T.; Lv, Z.; Fan, Y. Functional MRI Reveals Locomotion-Control Neural Circuits in Human Brainstem. Brain Sci. 2020, 10, 757.
- Bodurka, J.; Ye, F.; Petridou, N.; Murphy, K.; Bandettini, P.A. Mapping the MRI voxel volume in which thermal noise matches physiological noise--implications for fMRI. Neuroimage 2007, 34, 542–549.
- Evans, J.W.; Kundu, P.; Horovitz, S.G.; Bandettini, P.A. Separating slow BOLD from non-BOLD baseline drifts using multi-echo fMRI. Neuroimage 2015, 105, 189–197.
- Yan, L.; Zhuo, Y.; Ye, Y.; Xie, S.X.; An, J.; Aguirre, G.K.; Wang, J. Physiological origin of low-frequency drift in blood oxygen level dependent (BOLD) functional magnetic resonance imaging (fMRI). Magn. Reson. Med. 2009, 61, 819–827.
- Klein, A.; Andersson, J.; Ardekani, B.A.; Ashburner, J.; Avants, B.; Chiang, M.-C.; Christensen, G.E.; Collins, D.L.; Gee, J.; Hellier, P.; et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage 2009, 46, 786–802.
- Dadar, M.; Fonov, V.S.; Collins, D.L. Alzheimer’s Disease Neuroimaging Initiative. A comparison of publicly available linear MRI stereotaxic registration techniques. Neuroimage 2018, 174, 191–200.
- Napadow, V.; Dhond, R.; Kennedy, D.; Hui, K.K.; Makris, N. Automated brainstem co-registration (ABC) for MRI. Neuroimage 2006, 32, 1113–1119.
- Paxinos, G.; Huang, X.F. Atlas of the Human Brainstem, 1st ed.; Academic Press: San Diego, CA, USA, 1995.
- Eickhoff, S.B.; Stephan, K.E.; Mohlberg, H.; Grefkes, C.; Fink, G.R.; Amunts, K.; Zilles, K. A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. Neuroimage 2005, 25, 1325–1335.
- Oliva, V.; Gregory, R.; Davies, W.E.; Harrison, L.; Moran, R.; Pickering, A.E.; Brooks, J.C. Parallel cortical-brainstem pathways to attentional analgesia. Neuroimage 2020, 226, 117548.
- Brooks, J.C.; Beckmann, C.F.; Miller, K.L.; Wise, R.G.; Porro, C.A.; Tracey, I.; Jenkinson, M. Physiological noise modelling for spinal functional magnetic resonance imaging studies. Neuroimage 2008, 39, 680–692.
- Beissner, F.; Schumann, A.; Brunn, F.; Eisentrager, D.; Bar, K.J. Advances in functional magnetic resonance imaging of the human brainstem. Neuroimage 2014, 86, 91–98.
- Matt, E.; Fischmeister, F.P.S.; Amini, A.; Robinson, S.D.; Weber, A.; Foki, T.; Gizewski, E.R.; Beisteiner, R. Improving sensitivity, specificity, and reproducibility of individual brainstem activation. Brain Struct. Funct. 2019, 224, 2823–2838.
- Bao, R.; Wei, P.; Li, K.; Lu, J.; Zhao, C.; Wang, Y.; Zhang, T. Within-limb somatotopic organization in human SI and parietal operculum for the leg: An fMRI study. Brain Res. 2012, 1445, 30–39.
- Lambert, C.; Lutti, A.; Helms, G.; Frackowiak, R.; Ashburner, J. Multiparametric brainstem segmentation using a modified multivariate mixture of Gaussians. Neuroimage Clin. 2013, 2, 684–694.
- Lambert, C.; Chowdhury, R.; Fitzgerald, T.H.B.; Fleming, S.M.; Lutti, A.; Hutton, C.; Draganski, B.; Frackowiak, R.S.; Ashburner, J. Characterizing aging in the human brainstem using quantitative multimodal MRI analysis. Front. Hum. Neurosci. 2013, 7, 462.
- Zhu, J.; Wang, J.; Xu, C.; Zhang, X.; Qiao, L.; Wang, X.; Zhang, X.; Yan, X.; Ni, D.; Yu, T.; et al. The functional connectivity study on the brainstem-cortical/subcortical structures in responders following cervical vagus nerve stimulation. Int. J. Dev. Neurosci. 2020, 80, 679–686.
- Mills, E.P.; Akhter, R.; Di Pietro, F.; Murray, G.M.; Peck, C.C.; Macey, P.M.; Henderson, L.A. Altered Brainstem Pain Modulating Circuitry Functional Connectivity in Chronic Painful Temporomandibular Disorder. J. Pain. 2020.
- Mills, E.P.; Alshelh, Z.; Kosanovic, D.; Di Pietro, F.; Vickers, E.R.; Macey, P.M.; Henderson, L.A. Altered Brainstem Pain-Modulation Circuitry Connectivity during Spontaneous Pain Intensity Fluctuations. J. Pain Res. 2020, 13, 2223–2235.
- Caggiano, V.; Leiras, R.; Goñi-Erro, H.; Masini, D.; Bellardita, C.; Bouvier, J.; Caldeira, V.; Fisone, G.; Kiehn, O. Midbrain circuits that set locomotor speed and gait selection. Nature 2018, 553, 455–460.
- Ferreira-Pinto, M.J.; Ruder, L.; Capelli, P.; Arber, S. Connecting Circuits for Supraspinal Control of Locomotion. Neuron 2018, 100, 361–374.
- Li, X.; Chen, L.; Kutten, K.; Ceritoglu, C.; Li, Y.; Kang, N.; Hsu, J.T.; Qiao, Y.; Wei, H.; Liu, C.; et al. Multi-atlas tool for automated segmentation of brain gray matter nuclei and quantification of their magnetic susceptibility. Neuroimage 2019, 191, 337–349.
- Frangos, E.; Ellrich, J.; Komisaruk, B.R. Non-invasive Access to the Vagus Nerve Central Projections via Electrical Stimulation of the External Ear: fMRI Evidence in Humans. Brain Stimul. 2015, 8, 624–636.
- Sclocco, R.; Garcia, R.G.; Kettner, N.W.; Isenburg, K.; Fisher, H.P.; Hubbard, C.S.; Ay, I.; Polimeni, J.R.; Goldstein, J.; Makris, N.; et al. The influence of respiration on brainstem and cardiovagal response to auricular vagus nerve stimulation: A multimodal ultrahigh-field (7T) fMRI study. Brain Stimul. 2019, 12, 911–921.
- Cox, R.W.; Jesmanowicz, A. Real-time 3D image registration for functional MRI. Magn. Reson. Med. 1999, 42, 1014–1018.
- Eippert, F.; Kong, Y.; Winkler, A.M.; Andersson, J.L.; Finsterbusch, J.; Büchel, C.; Brooks, J.C.; Tracey, I. Investigating resting-state functional connectivity in the cervical spinal cord at 3T. Neuroimage 2017, 147, 589–601.
- Woods, R.P.; Grafton, S.T.; Holmes, C.J.; Cherry, S.R.; Mazziotta, J.C. Automated image registration: I. General methods and intrasubject, intramodality validation. J. Comput. Assist. Tomogr. 1998, 22, 139–152.
- Eickhoff, S.B.; Paus, T.; Caspers, S.; Grosbras, M.-H.; Evans, A.C.; Zilles, K.; Amunts, K. Assignment of functional activations to probabilistic cytoarchitectonic areas revisited. Neuroimage 2007, 36, 511–521.
- Fonov, V.; Le Troter, A.; Taso, M.; De Leener, B.; Lévêque, G.; Benhamou, M.; Sdika, M.; Benali, H.; Pradat, P.-F.; Collins, D.L.; et al. Framework for integrated MRI average of the spinal cord white and gray matter: The MNI-Poly-AMU template. Neuroimage 2014, 102, 817–827.
- De Leener, B.; Lévy, S.; Dupont, S.M.; Fonov, V.S.; Stikov, N.; Collins, D.L.; Callot, V.; Cohen-Adad, J. SCT: Spinal Cord Toolbox, an open-source software for processing spinal cord MRI data. Neuroimage 2017, 145, 24–43.
- Naidich, T.P.; Duvernoy, H.M.; Delman, B.N.; Sorensen, A.G.; Kollias, S.S.; Haacke, E.M. Duvernoy’s Atlas of the Human Brain Stem and Cerebellum, 1st ed.; Springer-Verlag/Wien: Vienna, Austria, 2009; pp. 63–89.
- Yang, G.; Zhou, S.; Bozek, J.; Dong, H.-M.; Han, M.; Zuo, X.-N.; Liu, H.; Gao, J.-H. Sample sizes and population differences in brain template construction. Neuroimage 2020, 206, 116318.
- Li, M.; Wang, D.; Ren, J.; Langs, G.; Stoecklein, S.; Brennan, B.P.; Lu, J.; Chen, H.; Liu, H. Performing group-level functional image analyses based on homologous functional regions mapped in individuals. PLoS Biol. 2019, 17, e2007032.
- Kong, R.; Li, J.; Orban, C.; Sabuncu, M.R.; Liu, H.; Schaefer, A.; Sun, N.; Zuo, X.-N.; Holmes, A.J.; Eickhoff, S.B.; et al. Spatial Topography of Individual-Specific Cortical Networks Predicts Human Cognition, Personality, and Emotion. Cereb. Cortex 2019, 29, 2533–2551.
