As the direct regulatory role of p53 and some of its isoform proteins are becoming established in modulating gene expression in cancer research, another aspect of this mode of gene regulation that has captured significant interest over the years is the mechanistic interplay between p53 and micro-RNA transcriptional regulation. The input of this into modulating gene expression for some of the cathepsin family members has been viewed as carrying noticeable importance based on their biological effects during normal cellular homeostasis and cancer progression.
- p53,cathepsin,micro-RNA,miRNA,cancer
1. Introduction
The tumor suppressor gene TP53 is mutated at a high frequency in a whole range of malignant diseases and has therefore been intensely researched for many years [1]. As is to be expected, the number of molecular networks that it has been shown to fundamentally regulate have also grown with great diversity and include aspects of DNA repair [2], cell senescence [3], angiogenesis [4], apoptosis [5][6] and cell cycle regulation [7]. While the main role of p53 in most of these processes are through its being able to directly regulate gene expression upon DNA binding, it can also mediate this through interacting with other transcription factors and regulators [8]. In some of its genetically mutated forms (mut-p53), p53 can take on the properties of a protein that is oncogenic, while some mutated derivatives can simply be inactive at the genetic or protein level [9]. Similarly, one key contributing factor originates from p53 being expressed as isoform proteins arising from the use of alternative promoters, translation initiation sites and mRNA splicing sites and which can act individually or in concert in modulating gene expression (Figure 1) [10][11].
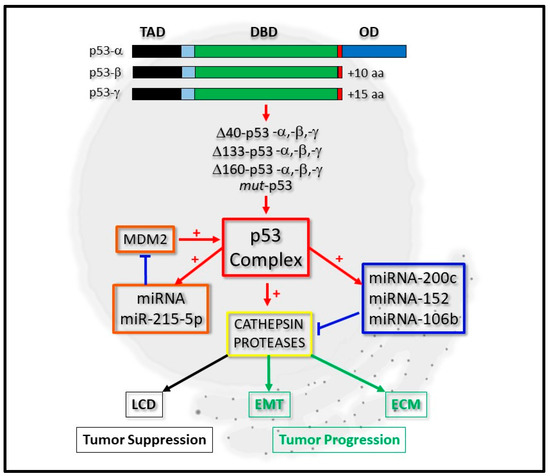
Figure 1. Integrative regulation of cathepsin proteases by p53 and micro-RNA expression. P53-alpha (p53-α) can be expressed as p53-beta (p53-β) or p53-gamma (p53-γ) isoform proteins, which lack the oligomerization domain (OD). Derivatives of these, which lack the complete Transactivation domain (TAD), but retain the DNA-binding domain (DBD), can also be expressed as Δ40-p53, Δ133-p53, Δ160-p53 or mut-p53 forms. The p53 complex can be regulated by micro-RNA (miRNA) expression through a positive feedback loop by positively regulating miRNA-215-5p, which negatively regulates MDM2 protein levels (orange boxes). It can transcriptionally regulate cathepsin protease expression directly or indirectly through directly regulating the expression of miRNA-200c, for example (blue box). Cathepsin protease expression (yellow box) contributes to lysosomal-mediated cell death (LCD) as a tumor suppressor (black boxes) or cell differentiation through Epithelial-Mesenchymal Transition (EMT) and the Extracellular Matrix (ECM) during tumor progression (green boxes).
From a regulatory perspective, p53 protein levels are kept to a minimum, through its polyubiquitination and destabilization by MDM2 and the proteasomal degradation pathway [9][10][11]. However, upon treating mammalian cells with oxidative stress or cytotoxic agents, nuclear p53 can become stabilized and modulate gene expression of proteins central to mediating cell arrest, DNA repair or apoptosis [12][13][14][15]. Additionally, post-translational modifications can also regulate p53 activity that mechanistically contribute to its cytoplasmic translocation, and where it can mediate mitochondria- or lysosomal-mediated cell death [16][17].
With over 14000 micro-RNAs annotated from the human genome that can regulate as much as 30% of all mRNAs expressed intracellularly, it is interesting to note that over 46% micro-RNA promoters have been reported to contain putative p53 binding sites [12][13]. While this highlights a potential direct link between p53 protein activation and micro-RNA expression, another important and direct role for the p53 protein in miRNA processing has also emerged. Here, p53 (or transcriptionally inactive p53) was revealed to be a central regulator of micro-RNA processing, through its ability to modulate the maturation of the micro-RNAs and their accessibility to mature mRNA messengers through its association with the protein Drosha [14] and the RISC complex (reviewed in [15][16]). Of importance is the ubiquitin ligase MDM2, which is under micro-RNA-mediated control as seen through the inhibitory actions of miRNA-192, miRNA-194, miRNA-215, miRNA-143, miRNA-145, and miRNA-605 expression [17]. For example, loss of miRNA-215-5p expression can enhance expression of MDM2, which results in diminished p53 protein levels [18]. As reported therein, p53 also positively regulated miRNA-215-5p expression, highlighting the existence of a p53 positive feedback loop [18]. Similarly, another good example of p53 regulation, through a miRNA acting on upstream activators of p53, occurs through miRNA-34, which acts by down-regulating the expression of the SIRT1 and HDAC intermediates that negatively-regulate p53 through its deacetylation (reviewed in [19]). While the actions of such micro-RNAs may give rise to enhanced levels of active p53 protein at the transcription and translation levels indirectly, p53 transcripts can also be directly targeted by miRNA-25 and miRNA-125b expression (reviewed in [16]).
The cathepsin proteases are a family of proteins that are developing greater importance due to them being intimately linked to tumor progression [20][21] and suppression [22]. During cancer progression, not only do they modulate the extracellular matrix and permit the dispersal of tumor cells following tumor growth, some of them also modulate the trans-differentiation of cells through the process of Epithelial Mesenchymal Transition (EMT) [21][23]. Simultaneously, the transcriptional regulation of cathepsins by p53 is also an area of research that is gaining much attention [22][24], particularly as lysosomes become more prone to lysis by lysosomorphic and cytotoxic agents upon cathepsin over-expression [25][26] and through p53 directly modulating lysosomal-mediated cell death [27][28], (Figure 1).
Consequently, the scientific interests revolving around the regulatory axis shared by all forms of p53, micro-RNAs and the cathepsins have captured the attention of many basic researchers, with a view to defining their co-regulatory relationships in greater depth (Figure 1).
2. The Biochemical Significance of the p53 Isoform Proteins
The p53 protein was first described over 30 years ago and its biological significance since then has had a significant amount of input into many of the p53-related paradigms that have been developed in many aspects of cancer cell biology. During this time, the TP53 gene has also revealed itself to encode a number of important p53 isoforms proteins [10][11], which have set many precedents while laying a number of very strong foundations for the characterization of the subsequently discovered p53 somatic mutations with relative ease [9]. For simplicity, the p53 isoforms can be categorized into two groups (Figure 1). The first group contains the p53-α, p53-β and p53-γ forms (which respectively encode WT-p53 (wild-type p53) and isoforms lacking the carboxyl-terminal Oligomerization Domain (OD), which is replaced with 10–15 amino acid extensions formed through alternative splicing of the mRNA (Figure 1 and Table 1). While these are driven transcriptionally from the promoter upstream of the first exon [29], Δ40-p53 isoform derivatives can also arise through the alternative splicing of the p53 transcript and the use of the initiator AUG at codon 40 [30]. Additional p53 protein derivatives (lacking part of its amino-terminal) can also arise from transcripts being driven from a second promoter located between intron 1 and exon 5, giving rise to ΔN-terminal p53 isoforms which have a 133 and 160 amino acid deletion at the amino-terminal [30][31][32]. Broadly, the p53 derivatives lacking the amino termini can be categorized into the second group (Figure 1 and Table 1).
Table 1. p53 isoform proteins.
|
p53 Isoform |
Amino Acids |
Protein (kD) |
Reference |
|
p53-α |
1-393 |
53 |
[33] |
|
p53-β |
1-331+10 |
47 |
[10] |
|
p53-γ |
1-331+15 |
48 |
[29] |
|
Δ40-p53-α |
40-393 |
47 |
|
|
Δ40-p53-β |
40-331+10 |
42 |
[29] |
|
Δ40-p53-γ |
40-331+15 |
42 |
[29] |
|
Δ133-p53-α |
133-393 |
35 |
[29] |
|
Δ133-p53-β |
133-331+10 |
29 |
[29] |
|
Δ133-p53-γ |
133-331+15 |
29 |
[29] |
|
Δ160-p53-α |
161-393 |
31 |
[32] |
|
Δ160-p53-β |
161-331+10 |
26 |
|
|
Δ160-p53-γ |
161-331+15 |
26 |
Biologically, all of the p53 isoforms exhibit diverse degrees of dominant-inhibitory effects for trans-activating gene expression through their abilities to form homo-tetramers or hetero-tetramers with WT-p53 [30][31][34]. This is based upon some of the isoforms lacking the OD, the full trans-activating domain (TAD) and showing varying degrees of protein stability and transcriptional activity based on the presence or absence of key phosphorylation sites, such as Ser-46 [35][36][37][38] and the carboxyl-terminal MDM2-specific ubiquitination sites [34][39]. Importantly, their biochemical characterization has indeed helped in offering an insight into how the p53 proteins arising from somatic mutations within the TP53 gene may differ biochemically in comparison to WT-p53 (or p53-α). Such mutations can be broadly described as a gain of function (GOF) or a loss of function (LOF) and the most commonest of them are the R175, G245, R248, R249, R273 and R282 mutants (collectively known as mut-p53) and which make up around 30% of all mutations found within the TP53 gene [40][41][42].
More specifically, the characterization of such p53 mutants has offered some excellent mechanistic insights into how certain micro-RNAs are regulated transcriptionally, especially in the context of cancer progression. For example, as far back as 2011, Chang et al. (2011) reported that miRNA-200c expression could be down-regulated upon the expression of a number of mut-p53 derivatives in 106 patient samples and MCF12A BC cells, which correlated significantly with tumor grade [43]. More recently, the expression of mut-p53 has also been linked to decreased miRNA-200c expression in human osteosarcoma cells by Tamura et al. (2015, [44]) and Alam et al. (2017, [45]) who identified the R280K mut-p53 protein as being responsible for this [45]. Here, increased expression levels of Moesin in MCF7 1001 BC cells were reported, as a significant contributing factor to carcinogenesis.
Collectively, the existence of such a high number of p53 isoform proteins can potentially offer a number of alternative mechanisms for how the TP53 gene can exert its biological effects. Consequently, their importance in being able to regulate tumor suppressive miRNA expression, either exclusively or with WT-p53, is being viewed as mechanistically significant during tumor initiation or progression.
3. p53, micro-RNA Regulation and Cathepsin Proteases: A Developing Network
The family of cathepsin proteases is composed of aspartate proteases (D, E), serine proteases (A, G) and the cysteine proteases (B, C, F, H, K, L, O, S, V, Z/X, W) [20]. Collectively, they are expressed as inactive zymogens, which have the capability to become auto-activated or trans-activated as they traffic from the endosome to reside within the lysosome, but can also be found in the nucleus [46]. Some of them are upregulated in expression, especially during cancer progression and can be secreted into the Extracellular Matrix (ECM) where they can modulate ECM components and contribute to malignancy [47][48]. Nevertheless, normally they are localized within the lysosome, from where they can leak into the cytoplasm and activate intermediates from the intrinsic apoptotic pathway as in the case of BID cleavage, causing the activation of apoptosis [49].
More recently, cathepsins L and D have been seen to reside in the nucleus where they can cleave the Histone H3 protein [50][51], CUX1 [52][53][54], TRPS1 [23] and enhance proliferation, induce EMT and increase the motility of cells. Consequently, a strong interest in how cathepsin expression is regulated has developed with the transcriptional regulation of cathepsins D and L having been linked to p53. Here, cathepsin D was expressed in a p53-dependent manner in U1752, Pa1 and ML1 leukemia cell manner and p53 was reported to bind to two p53 consensus sequences within the cathepsin D promoter [22]. Similarly, p53 could bind the promoter region of cathepsin L and the expression of which could also be driven by mut-p53 expression in glioblastoma cells [24]. Being mindful of these observations, there are justifiable reasons for why the scope of research here needs to be broadened in order to ascertain how cathepsins may be regulated in the absence and presence of p53 (or its isoforms and mut-p53 derivatives), and whether such events can still permit the cathepsins to drive tumor progression.
Generally speaking, developing interests have revolved around how cathepsin genes may be regulated by specific micro-RNAs, and of importance here is how these may be linked to what is commonly known about p53 and cathepsin protease regulation. In the instance of cathepsin proteases, this area of research appears to be relatively undeveloped, and being mindful of there being around 15 cathepsin proteases (with the majority of them being linked to cancer development or progression [20]), reportedly only a few of them appear to be regulated by micro-RNAs that have a direct or indirect connection with p53. Moreover, the regulation of cathepsins in the context of p53 isoforms or mutant-derivatives thereof appear to be even less explored and is an important consideration in light of how quickly this area of p53 biology is expanding.
Consequently, in highlighting the nature of these developing integrative regulatory networks, the next section is devoted to reviewing, a) which micro-RNAs are regulated by (or regulate) p53, and b) how these micro-RNAs regulate cathepsin protease family members in the context of cancer, with a view to broadening our understanding of the regulatory interplay between p53 and cathepsin transcription. Broadly speaking, miRNA-200c, miRNA-152 and miRNA-106b appear to be the most characterized in this context, with others such as miRNA-29a (cathepsin K, [55]), miRNA-30 (cathepsin D, [56]), miRNA-25-3p (cathepsin K, [57]), miRNA-140 (cathepsin B, [58]), miRNA-483-5p (cathepsin K, [59]) and miRNA-506-3p (cathepsin K, [60]) being characterized to a lesser extent (Table 2).
Table 2. The developing networks between micro-RNA, cathepsin proteases and p53 expression.
|
Micro-RNA |
Cathepsin |
p53 isoform |
Cell Type |
Reference |
|
miRNA-200c |
L |
WT-p53-α |
A549 Lung |
|
|
miRNA-152 |
L |
WT-p53-α |
Gastrointestinal |
|
|
miRNA-106b |
A |
WT-p53-α |
Colorectal |
|
|
miRNA-140 |
B |
- |
Glioblastoma |
[58] |
|
miRNA-30 |
D |
- |
Macrophage |
[56] |
|
miRNA-25-3p |
K |
- |
Osteoblast |
[57] |
|
miRNA-483-5p |
K |
- |
PBMC |
[59] |
|
miRNA-506-3p |
K |
- |
Macrophage |
[60] |
|
miRNA-29a |
K |
- |
Osteoblast |
[55] |
The expression of micro-RNAs connected with cathepsin gene expression are highlighted in conjunction with specific p53 isoforms and cell types they have been collectively characterized in (WT-p53, wild-type p53; PBMC, Peripheral Blood Mononuclear Cells; -, unknown).
4. miRNA-200c, -152, -106b Expression and Cancer Progression: A Clinical Perspective
Based on the above, there are clear regulatory relationships that are emerging between p53, cathepsin and micro-RNA expression. While the focus of this article has so far been originated from defining the molecular roles that p53 and cathepsins share in disease progression, for completeness we would like to extend the importance of the above miRNAs within a clinical context. This has great significance through the common biological traits their downstream target gene products share with some of the cathepsin proteases, and therefore it is worth focusing on this through highlighting alternative transcripts (or proteins) that are targeted by these miRNAs. In addition to this, we would like to review the recent progress on how these micro-RNAs are being utilized in diagnostic and prognostic assays. For simplicity and consistency, we will keep the focus and the context as close to lung cancer (miRNA-200c), gastric cancer (miRNA-152) and colorectal cancer (miRNA-106b), as possible.
4.1. miRNA-200c Expression
As far back as 2013, the importance of miRNA-200c in the regulation of disease progression has positively been gaining greater momentum. For example, the loss of miRNA-200c within the lungs [66] was seen to correlate with NSCLC cells showing an invasive and chemo-resistant phenotype [67], while positive expression of it could sensitize cells to chemotherapeutic [68] and radiotherapeutic [69] agents. As reported by Cortez et al. (2014), such findings could be extended and they reported the expression of miRNA-200c enhanced radio-sensitivity of cells in a xenograft lung cancer model through miRNA-200c expression inducing the oxidative stress response by its regulation of oxidative response genes [70]. Similarly, Shi et al. (2013) showed that A549 cells could be radio-sensitized upon miRNA-200c expression [69], while Kopp et al. (2013) showed that miRNA-200c could target K-Ras expression and that it could inhibit tumor progression and therapeutic resistance in a panel of BC cell lines [71]. Additional tumor suppressive effects have also been reported and which showed miRNA-200c expression to decrease NCCLC and A549 migration or invasiveness. MiRNA-200c was also reported to target USP25 [72], ZEB1 [73] or ZEB2 [74], and had the effect of modulating cell migration and differentiation of cells. Similarly, miRNA-200c expression was also correlated with reduced cell migration of H23 cells through enhanced E-cadherin expression [75]. Conversely, miRNA-200c was also seen to function by inducing cell death through the apoptotic pathway. For example, Bai et al. (2014) showed that miRNA-200c expression targeted the RECK gene and induced the apoptotic death of H460 lung cells, which was enhanced in the presence of Reservatol stimulation [76]. Generally, the functional role of positive miRNA-200c expression appears to be one that minimizes tumor progression and is mechanistically linked to the suppression of genes that have an oncogenic effect (Table 3).
Table 3. Elevated (+) or reduced (−) miRNA-200c levels are shown, as are their target genes, their biological effects and whether these factors can sensitize cells to certain therapeutic agents. The cell types indicate the types of cells characterized. BC, breast cancer; NSCLC, non-small cell lung cancer.
|
micro-RNA |
Target |
Negative Effect |
Sensitizing Agent |
Cell Type |
Reference |
|
200c (+) |
VEGF, VEGFR2 |
Angiogenesis, Cell Migration |
Radiation |
A549 |
[69] |
|
200c (+) |
PRDX2, SENS1, GABPA/Nrf2 |
Oxidative Response |
Radiation |
A549, H460, H1299 |
[70] |
|
200c (+) |
K-Ras |
Proliferation, Cell cycle |
− |
Lung and BC cell lines |
[71] |
|
200c (+) |
USP25 |
Cell Migration EMT |
− |
NSCLC cell lines |
[72] |
|
200c (−) |
ZEB1 |
Cell Migration |
Gefitinib |
PC-9-ZD |
[73] |
|
200c (−) |
ZEB2 |
EMT |
− |
A-549 |
[74] |
|
200c (+) |
Possibly E-cadherin |
Cell Migration |
− |
H23, A549, HCC-44 |
[75] |
|
200c (+) |
Possibly RECK |
Proliferation |
Reservatol |
H-460 |
[76] |
Simultaneously, a number of excellent studies have also published how miRNA evaluation in cells can be successfully utilized as a diagnostic and prognostic tool. For example, Tejero et al. (2014) reported that miRNA-200c could be a good biomarker for overall survival (OS) during the early stages of NSCLC adenocarcinoma [75]. Here, qRT-PCR was used to evaluate 155 resected patient tumor samples for miRNA-200c expression and their findings complimented with functional studies using H23, HCC44 and A549 cell lines. Elevated miRNA-200c expression in early stage NSCLC was significantly correlated with a decrease in OS [75]. Similarly, Kim et al. (2014) reported miRNA-200c expression to be significantly up-regulated and correlated with tumor size, lymphovascular invasion and poor OS [77]. Other publications supporting such trends have also been recently reported through the extensive use of meta-analyses to help define the diagnostic potential of miRNA-200c expression. For example, Shao et al. (2015) correlated high levels of circulating miRNA-200c with a poor OS and PFS (in advanced disease) and low miRNA-200c levels with poor survival during early stages of disease [78]. Here, 18 published studies were analyzed and the regulation of EMT (or MET) by miRNA-200c was seen as a possible cause. Teng et al. (2016) identified circulating and tissue-derived miRNA-200c as a potential diagnostic and prognostic marker for epithelial ovarian cancer (EOC) [79]. Si et al. (2017) analyzed 110 resected tumor samples from NSCLC patients for quantification of miRNA-200c, the expression of which was associated with positive lymph node metastasis, TNM classification and a reduced 5 year disease-free survival rate [80]. More recently, the use of miRNA as biomarkers for responsiveness to chemotherapeutics have also gained some attention as reported by Li et al. (2017). Here, the findings from 46 published articles showed that low expression levels of miRNA-200c (or IHC negative staining) was a good predictor for responsiveness to chemo- or radio-therapy in esophageal cancer [81]. Moreover, Zheng et al. (2017) used a meta-analysis from 60 reported studies to highlight that increased miRNA-200c expression correlated with poor prognosis in gastrointestinal cancer (GIC) patients [82], while increased miRNA-200c expression offered a better OS for ovarian cancer (OC) patients, as reported by Shi et al. (2018) [83], (Table 4).
Table 4. Elevated (+) or reduced (−) miRNA-200c levels are shown, as are the cancer types, source of materials the miRNA was detected from and the patient cohort size. NSCLC, non-small cell lung cancer; EOC, epithelial ovarian cancer; GIC, gastrointestinal cancer; esophageal cancer (ES); OC, ovarian cancer. The negative or positive use of the technique in diagnostic or prognostic evaluation of patients are denoted by − or +, respectively.
|
micro-RNA |
Cancer type |
Source |
Cohort Size |
Diagnostic |
Prognosis |
Reference |
|
200c (+) |
NSCLC |
Tissue |
155 |
− |
Reduced |
[75] |
|
200c (+) |
NSCLC |
Tissue |
72 |
− |
Reduced |
[77] |
|
200c (−) |
varied |
Tissue/Blood |
18 studies |
− |
Poor OS and PFS |
[78] |
|
200c (+) |
EOC |
Tissue/Plasma |
14 studies |
+ |
+ |
[79] |
|
200c (+) |
NSCLC |
Tissue |
110 |
− |
Reduced |
[80] |
|
200c (−) |
EC |
Tissue |
46 studies |
− |
+ |
[81] |
|
200c (+/−) |
GIC |
Tissue/Blood |
60 studies |
− |
+ |
[82] |
|
200c (+) |
OC |
Tissue/Blood |
15 studies |
− |
+ |
[83] |
4.2. miRNA-152 Expression
The expression of miRNA-152 has been evaluated in a number of cancers associated with the gastrointestinal tract over the last 10 years with some very clear findings on which target genes may be regulated by miRNA-152 and what role they may play during cancer progression. For example, Chen et al. (2010) analyzed 101 gastric cancer (GC) and colorectal cancer (CRC) tissue samples and reported a decrease in miRNA-152 expression, which correlated with an increased tumor size and advanced pT stage in GIC, and inversely correlated with cholecystokinin B receptor protein expression in GC [84]. Other target genes for miRNA-152 include PIK3CA in breast cancer (BC) [85] or PIK3R3 in CRC [86], EPAS1 in Paclitaxel-resistant BC cells [87], CD151 in GC [88], IGF-1R and IRS1 in BC [89], B7-H1 in GC [90], CDK8 in hepatocellular carcinoma (HCC) [91], p27 in bone marrow cells [92], SOS1 in Glioblastoma (GBM) [93] cells and KLF4 in colon cancer (CC) cells [94], (Table 5).
Table 5. Elevated (+) or reduced (−) miRNA-152 levels are shown, as are their target genes, their biological effects and whether these factors can sensitize cells to certain therapeutic agents. The cell types indicate the types of cells characterized. BC, breast cancer; GC, gastric cancer; CRC, colorectal cancer; BM, bone marrow; GBM, glioblastoma; HCC, hepatocellular carcinoma; CC, colon cancer.
|
micro-RNA |
Target |
Negative Effect |
Sensitizing Agent |
Cell Type |
Reference |
|
152 (−) |
PIK3CA |
Cell Proliferation |
− |
HCC1806 |
[85] |
|
152 (−) |
PIK3R3 |
Cell Proliferation Migration |
− |
CRC cell lines |
[86] |
|
152 (−) |
EPAS |
Apoptosis |
Paclitaxel |
BC cell lines |
[87] |
|
152 (−) |
CD151 |
Proliferation Migration |
− |
GC Tissues |
[88] |
|
152 (−) |
IGF-1R |
Proliferation Angiogenesis |
− |
BC Tissues |
[89] |
|
152 (−) |
IRS1 |
Proliferation Angiogenesis |
− |
BC Tissues |
[89] |
|
152 (−) |
B7-H1 |
T-cell Proliferation |
− |
GC cell lines |
[90] |
|
152 (−) |
CDK8 |
Proliferation Apoptosis |
− |
HCC cell lines |
[91] |
|
152 (+) |
p27 |
Proliferation |
− |
BM cells, K562 |
[92] |
|
152 (−) |
SOS1 |
Proliferation Apoptosis |
Cisplatin |
GBM cell lines |
[93] |
|
152 (+) |
KLF4 |
Proliferation |
− |
CC cell lines |
[94] |
At the clinical level, Safrinzo et al. (2013) showed stage I-IIIA NSCLC patient plasma samples to contain decreased miRNA-152 expression levels, which correlated with decreased DFS for lung squamous cell carcinoma prevalence (SCC) [95]. Li et al. (2016) reported a decrease in expression of miRNA-152 in CRC tissues which inversely correlated with TNM staging and lymph node metastases [86], while Wang et al. (2017) observed a decrease in miRNA-152 expression in GC patients [90] and Ge et al. (2017) showed that miRNA-152-3p could target PIK3CA in BC as a tumor suppressor [85]. You et al. (2018) analyzed 15 GC tissues and confirmed that miRNA-152-3p expression was reduced and could directly target PIK3CA in SGC-7901 cells [96]. Alternatively, Matin et al. (2018) profiled 372 patient plasma samples collected before, during and after treatments for PC and elevated miRNA-152-3p levels reported, while (interestingly) low levels of miRNA-152-3p expression were observed in prostate cancer (PC) samples [97]. Such findings indeed highlight the power of miRNA-152 quantification as a diagnostic marker for PC (as seen for lung cancer, CRC and BC [98]). In CML, miRNA-152-3p expression was elevated in bone marrow (BM) samples and upon expression of miRNA-152-3p in K562 cells, proliferation was decreased and apoptosis levels were enhanced through targeting the p27 (CDKN1B) gene [92]. From the analysis of 89 HCC tumor samples, Yin et al. (2019) showed that miRNA-152-3p levels were decreased and which correlated with tumor volume and TNM staging [91]. Moreover, Wang et al. (2017) saw that decreased miRNA-152 expression was related to poor OS and DFS in GC, which could be used as an independent risk factor for the prediction of HCC prognosis [90]. More recently, Li et al. (2019) diagnosed early stage I-II BC by screening 106 plasma samples and tissues for miRNA-152-3p expression and reported it to be decreased, which correlated with ER-positive and PR-positive patients [99]. Finally, Song et al. (2020) observed reduced levels of miRNA-152-3p in a study of 30 invasive BC samples, which correlated with a poor prognosis [87] and the overexpression of which could sensitize chemo-resistant BC cells to Paclitaxel-mediated cell death (Table 6).
Table 6. Elevated (+) or reduced (−) miRNA-152 levels are shown, as are the cancer types, source of materials the miRNA was detected from and the patient cohort size. The negative or positive use of the technique in diagnostic or prognostic evaluation of patients are denoted by - or +, respectively. NSCLC, non-small lung cancer cells; CRC, colorectal cancer; PC, prostate cancer; BC, breast cancer; GC, gastric cancer; CML, chronic myelogenous leukemia; HCC, hepatocellular carcinoma.
|
micro-RNA |
Cancer type |
Source |
Cohort size |
Diagnostic |
Prognosis |
Reference |
|
152 (−) |
CRC |
Tissue |
28 |
+/− |
− |
[86] |
|
152 (−) |
BC invasive |
Tissue |
30 |
− |
Poor |
[87] |
|
152 (−) |
GC |
Tissues |
42 |
− |
− |
[90] |
|
152 (+) |
CML |
Bone Marrow |
40 |
- |
- |
[90] |
|
152 (−) |
HCC |
Tissue |
89 |
− |
+/− |
[91] |
|
152 (−) |
Stage I-IIIA NSCLC |
Plasma |
52 |
− |
Reduced DFS |
[95] |
|
152 (−) |
PC, lung, CRC, BC |
Plasma |
204 |
+ |
− |
[98] |
|
152 (−) |
BC stage I-II |
Plasma |
106 |
+ |
− |
[99] |
4.3. miRNA-106b Expression
While miRNA-106b expression has indeed emerged as having a tangible biological effect in most cancer cell systems, the outcomes from such studies at this moment have offered mixed results and appears to be an area of research development. Cai et al. (2011) reported that miRNA-106b could target RB expression in laryngeal carcinoma [100] and ATG16L1 expression in Crohn’s Disease samples [101][102]. Additionally, all three micro-RNAs from the miRNA-106b-25 cluster were seen to target PTEN expression [103][104] and increased miRNA-106b expression recorded in CRC tissues which could target DLC-1 (while enhancing EMT, [105]) and FAT4 in CRC tissues or cell lines [106] (Table 7).
Table 7. Elevated (+) miRNA-106b levels are shown, as are their target genes, their biological effects and whether these factors can sensitize cells to certain therapeutic agents. The cell types indicate the types of cells characterized. CD, Crohn’s Disease; CRC, colorectal cancer.
|
micro-RNA |
Target |
Positive Effect |
Sensitizing Agent |
Cell Type |
Reference |
|
106b (+) |
RB |
Reduced Cell Arrest |
− |
Laryngeal carcinoma HEP2G+T1U212 |
[100] |
|
106b (+) |
ATG16L1 |
Decreased Autophagy |
− |
CD |
|
|
106b (+) |
PTEN |
Tumor Initiation Stemness |
Radiation |
CRC cell lines |
[104] |
|
106b (+) |
p21 (indirectly) |
Tumor Initiation Stemness |
Radiation |
CRC cell lines |
[104] |
|
106b (+) |
DLC-1 |
EMT |
− |
CRC Tissues CRC cell lines |
[105] |
|
106b (+) |
FAT4 |
Viability Angiogenesis Migration |
− |
CRC Tissues CRC cell lines |
[106] |
Based on the growing importance of utilizing miRNA expression within the clinic, their quantification for the diagnosis and prognosis of patients has moved in a positive direction. In the instance of miRNA-106b a number of excellent studies have significantly shaped this area and are worth mentioning.
As far back as 2010, Wang et al. (2010) analyzed CRC samples using qRT-PCR and found miRNA-106b to be up-regulated [107] as confirmed thereafter in colorectal cancer stromal tissues as well [108]. Subsequently, Wang et al. (2015) found miRNA-106b expression to be increased in 180 CRC patients, which correlated with a longer OS but were not seen as being statistically significant [109]. Similarly, Zhang et al. (2015) analyzed 95 CRC patient samples and miRNA-106b expression correlated with a shorter OS or DFS and which had significant reliability as an independent prognostic factor for CRC [110]. In the context of RCCC, Gu et al. (2015) performed a meta-analysis on 27 studies analyzing the expression of miRNA-106b, and (unlike CRC) reported that a decreased miRNA-106b was associated with a poor prognosis [111]. More recently, high exosomal miRNA-106b levels from the serum have been reported to correlate with a high TNM stage, a larger tumor volume and a poor prognosis [105].
Collectively, such findings support the notion that the use of miRNA-106b as a prognostic marker is unreliable, based on inconsistencies reported from a number of studies correlating miRNA expression levels with tumor grade (Table 8).
Table 8. Elevated (+) or reduced (−) miRNA-106b levels are shown, as are the cancer types, source of materials the miRNA was detected from and the patient cohort size. The negative or positive use of the technique in diagnostic or prognostic evaluation of patients are denoted by − or +, respectively. Exo, exosomal; RCCC, renal clear cell carcinoma; CC, colon cancer; CRC, colorectal cancer; OS, overall survival, DFS, disease-free survival; *, not statistically significant.
In summary, relatively good progress is being made in defining target genes for the above specific miRNAs, which may help to offer a broader perspective on how other genes of importance may synergize with cathepsin regulation in disease progression. Moreover, additional insights are also emerging into how such micro-RNAs can be utilized as reliable diagnostic and prognostic markers to possibly compliment on-going efforts with other biomarkers of importance, such as p53 and cathepsin expression. Additionally, from the above studies, oncogenic micro-RNAs are also emerging to play an important regulatory role in disease progression, and do have the potential to be targeted for therapeutic purposes using small molecule-inhibitors or -degraders (as reviewed in [112]) or through targeting specific upstream transcription regulatory signaling pathways.
This entry is adapted from the peer-reviewed paper 10.3390/cancers12113454
References
- Meek, D.W. Regulation of the p53 response and its relationship to cancer. Biochem. J. 2015, 469, 325–346, doi:10.1042/BJ20150517. Available online: https://www.ncbi.nlm.nih.gov/pubmed/26205489 (accessed on 5 October 2020).
- Williams, B.A.; Schumacher, B. P53 in the DNA-damage-repair process. Cold Spring Harb. Perspect. Med. 2016, 6, doi:10.1101/cshperspect.a026070. Available online: https://www.ncbi.nlm.nih.gov/pubmed/27048304 (accessed on 5 October 2020).
- Nicolai, S.; Rossi, A.; Di Daniele, N.; Melino, G.; Annicchiarico-Petruzzelli, M. DNA repair and aging: The impact of the p53 family. Aging 2015, 7, 1050–1065, doi:10.18632/aging.100858. Available online: https://www.ncbi.nlm.nih.gov/pubmed/26668111 (accessed on 5 October 2020)
- Pfaff, M.J.; Mukhopadhyay, S.; Hoofnagle, M.; Chabasse, C.; Sarkar, R. Tumor suppressor protein p53 negatively regulates ischemia-induced angiogenesis and arteriogenesis. J. Vasc. Surg. 2018, 68, 222S–233S, doi:10.1016/j.jvs.2018.02.055. Available online: https://www.ncbi.nlm.nih.gov/pubmed/30126780 (accessed on 5 October 2020).
- Ranjan, A.; Iwakuma, T. Non-canonical cell death induced by p53. Int. J. Mol. Sci 2016, 17, doi:10.3390/ijms17122068. Available online: https://www.ncbi.nlm.nih.gov/pubmed/27941671 (accessed on 5 October 2020).
- Wang, X.; Simpson, E.R.; Brown, K.A. P53: Protection against tumor growth beyond effects on cell cycle and apoptosis. Cancer Res. 2015, 75, 5001–5007, doi:10.1158/0008-5472.CAN-15-0563. Available online: https://www.ncbi.nlm.nih.gov/pubmed/26573797 (accessed on 5 October 2020).
- Chen, J. The cell-cycle arrest and apoptotic functions of p53 in tumor initiation and progression. Cold Spring Harb. Perspect. Med. 2016, 6, doi:10.1101/cshperspect.a026104. Available online: https://www.ncbi.nlm.nih.gov/pubmed/26931810 (accessed on 5 October 2020).
- Avantaggiati, M.L.; Ogryzko, V.; Gardner, K.; Giordano, A.; Levine, A.S.; Kelly, K. Recruitment of p300/cbp in p53-dependent signal pathways. Cell 1997, 89, 1175–1184, doi:10.1016/s0092-8674(00)80304-9. Available online: https://www.ncbi.nlm.nih.gov/pubmed/9215639 (accessed on 5 October 2020).
- Olivier, M.; Hollstein, M.; Hainaut, P. Tp53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010, 2, doi:10.1101/cshperspect.a001008. Available online: https://www.ncbi.nlm.nih.gov/pubmed/20182602 (accessed on 5 October 2020).
- Flaman, J.M.; Waridel, F.; Estreicher, A.; Vannier, A.; Limacher, J.M.; Gilbert, D.; Iggo, R.; Frebourg, T. The human tumour suppressor gene p53 is alternatively spliced in normal cells. Oncogene 1996, 12, 813–818. Available online: https://www.ncbi.nlm.nih.gov/pubmed/8632903 (accessed on 5 October 2020).
- Marcel, V.; Dichtel-Danjoy, M.L.; Sagne, C.; Hafsi, H.; Ma, D.; Ortiz-Cuaran, S.; Olivier, M.; Hall, J. Biological functions of p53 isoforms through evolution: Lessons from animal and cellular models. Cell Death Differ. 2011, 18, 1815–1824, doi:10.1038/cdd.2011.120. Available online: https://www.ncbi.nlm.nih.gov/pubmed/21941372 (accessed on 5 October 2020).
- Xi, Y.; Shalgi, R.; Fodstad, O.; Pilpel, Y.; Ju, J. Differentially regulated micro-rnas and actively translated messenger rna transcripts by tumor suppressor p53 in colon cancer. Clin. Cancer Res. 2006, 12, 2014–2024, doi:10.1158/1078-0432.CCR-05-1853. Available online: https://www.ncbi.nlm.nih.gov/pubmed/16609010 (accessed on 5 October 2020).
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; Da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; Rajewsky, N. Combinatorial microrna target predictions. Nat. Genet. 2005, 37, 495–500, doi:10.1038/ng1536. Available online: https://www.ncbi.nlm.nih.gov/pubmed/15806104 (accessed on 5 October 2020).
- Suzuki, H.I.; Yamagata, K.; Sugimoto, K.; Iwamoto, T.; Kato, S.; Miyazono, K. Modulation of microrna processing by p53. Nature 2009, 460, 529–533, doi:10.1038/nature08199. Available online: https://www.ncbi.nlm.nih.gov/pubmed/19626115 (accessed on 5 October 2020).
- Goeman, F.; Strano, S.; Blandino, G. Micrornas as key effectors in the p53 network. Int. Rev. Cell Mol. Biol. 2017, 333, 51–90, doi:10.1016/bs.ircmb.2017.04.003. Available online: https://www.ncbi.nlm.nih.gov/pubmed/28729028 (accessed on 5 October 2020).
- Hermeking, H. Micrornas in the p53 network: Micromanagement of tumour suppression. Nat. Rev. Cancer 2012, 12, 613–626, doi:10.1038/nrc3318. Available online: https://www.ncbi.nlm.nih.gov/pubmed/22898542 (accessed on 5 October 2020).
- Liu, J.; Zhang, C.; Zhao, Y.; Feng, Z. Microrna control of p53. J. Cell Biochem. 2017, 118, 7–14, 10.1002/jcb.25609.Available online: https://www.ncbi.nlm.nih.gov/pubmed/27216701 (accessed on 5 October 2020).
- Singh, A.; Bhattacharyya, N.; Srivastava, A.; Pruett, N.; Ripley, R.T.; Schrump, D.S.; Hoang, C.D. Microrna-215-5p treatment suppresses mesothelioma progression via the mdm2-p53-signaling axis. Mol. Ther. 2019, 27, 1665–1680, doi:10.1016/j.ymthe.2019.05.020. Available online: https://www.ncbi.nlm.nih.gov/pubmed/31227395 (accessed on 5 October 2020).
- Ong, A.L.C.; Ramasamy, T.S. Role of sirtuin1-p53 regulatory axis in aging, cancer and cellular reprogramming. Ageing Res. Rev. 2018, 43, 64–80, doi:10.1016/j.arr.2018.02.004. Available online: https://www.ncbi.nlm.nih.gov/pubmed/29476819 (accessed on 5 October 2020).
- Soond, S.M.; Kozhevnikova, M.V.; Zamyatnin, A.A., Jr. Patchiness' and basic cancer research: Unravelling the proteases. Cell Cycle 2019, 18, 1687–1701, doi:10.1080/15384101.2019.1632639. Available online: https://www.ncbi.nlm.nih.gov/pubmed/31213124 (accessed on 5 October 2020).
- Soond, S.M.; Kozhevnikova, M.V.; Townsend, P.A.; Zamyatnin, J.A.A. Cysteine Cathepsin Protease Inhibition: An update on its Diagnostic, Prognostic and Therapeutic Potential in Cancer. Pharmaceuticals 2019, 12, 87, doi:10.3390/ph12020087. Available online: https://www.ncbi.nlm.nih.gov/pubmed/31212661 (accessed on 5 October 2020).
- Wu, G.S.; Saftig, P.; Peters, C.; El-Deiry, W.S. Potential role for Cathepsin D in p53-dependent tumor suppression and chemosensitivity. Oncogene 1998, 16, 2177–2183, doi:10.1038/sj.onc.1201755. Available online: https://www.ncbi.nlm.nih.gov/pubmed/9619826 (accessed on 5 October 2020).
- Bach, A.-S.; Derocq, D.; Laurent-Matha, V.; Montcourrier, P.; Sebti, S.S.; Orsetti, B.; Theillet, C.; Gongora, C.; Pattingre, S.; Ibing, E.; et al. Nuclear cathepsin D enhances TRPS1 transcriptional repressor function to regulate cell cycle progression and transformation in human breast cancer cells. Oncotarget 2015, 6, 28084–28103, doi:10.18632/oncotarget.4394. Available online: https://www.ncbi.nlm.nih.gov/pubmed/26183398 (accessed on 5 October 2020).
- Katara, R.; Mir, R.A.; Shukla, A.A.; Tiwari, A.; Singh, N.; Chauhan, S.S. Wild type p53-dependent transcriptional upregulation of cathepsin L expression is mediated by C/EBPα in human glioblastoma cells. Biol. Chem. 2010, 391, 1031–40, doi:10.1515/bc.2010.103. Available online: https://www.ncbi.nlm.nih.gov/pubmed/20536385 (accessed on 5 October 2020).
- Ono, K.; Kim, S.O.; Han, J. Susceptibility of Lysosomes to Rupture Is a Determinant for Plasma Membrane Disruption in Tumor Necrosis Factor Alpha-Induced Cell Death. Mol. Cell. Biol. 2003, 23, 665–676, doi:10.1128/mcb.23.2.665-676.2003. Available online: https://www.ncbi.nlm.nih.gov/pubmed/12509464 (accessed on 5 October 2020).
- Fehrenbacher, N.; Bastholm, L.; Kirkegaard-Sørensen, T.; Rafn, B.; Bøttzauw, T.; Nielsen, C.; Weber, E.; Shirasawa, S.; Kallunki, T.; Jäättelä, M. Sensitization to the Lysosomal Cell Death Pathway by Oncogene-Induced Down-regulation of Lysosome-Associated Membrane Proteins 1 and 2. Cancer Res. 2008, 68, 6623–6633, doi:10.1158/0008-5472.can-08-0463. Available online: https://www.ncbi.nlm.nih.gov/pubmed/18701486 (accessed on 5 October 2020).
- Li, N.; Zheng, Y.; Chen, W.; Wang, C.; Liu, X.; He, W.; Xu, H.; Cao, X. Adaptor Protein LAPF Recruits Phosphorylated p53 to Lysosomes and Triggers Lysosomal Destabilization in Apoptosis. Cancer Res. 2007, 67, 11176–11185, doi:10.1158/0008-5472.can-07-2333. Available online: https://www.ncbi.nlm.nih.gov/pubmed/18056442 (accessed on 5 October 2020).
- Boya, P.; Andreau, K.; Poncet, D.; Zamzami, N.; Perfettini, J.-L.; Metivier, D.; Ojcius, D.M.; JäätteläM.; Kroemer, G. Lysosomal Membrane Permeabilization Induces Cell Death in a Mitochondrion-dependent Fashion. J. Exp. Med. 2003, 197, 1323–1334, doi:10.1084/jem.20021952. Available online: https://www.ncbi.nlm.nih.gov/pubmed/12756268 (accessed on 5 October 2020).
- Bourdon, J.-C.; Fernandes, K.; Murray-Zmijewski, F.; Liu, G.; Diot, A.; Xirodimas, D.P.; Saville, M.K.; Lane, D.P. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005, 19, 2122–2137, doi:10.1101/gad.1339905. Available online: https://www.ncbi.nlm.nih.gov/pubmed/16131611 (accessed on 5 October 2020).
- Courtois, S.; Verhaegh, G.; North, S.; Luciani, M.-G.; Lassus, P.; Hibner, U.; Oren, M.; Hainaut, P. ΔN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene 2002, 21, 6722–6728, doi:10.1038/sj.onc.1205874. Available online:https://www.ncbi.nlm.nih.gov/pubmed/12360399 (accessed on 5 October 2020).
- Ghosh, A.; Stewart, D.; Matlashewski, G. Regulation of Human p53 Activity and Cell Localization by Alternative Splicing. Mol. Cell. Biol. 2004, 24, 7987–7997, doi:10.1128/mcb.24.18.7987-7997.2004. Available online: https://www.ncbi.nlm.nih.gov/pubmed/15340061 (accessed on 5 October 2020).
- Marcel, V.; Perrier, S.; Aoubala, M.; Ageorges, S.; Groves, M.J.; Diot, A.; Fernandes, K.; Tauro, S.; Bourdon, J.-C. Δ160p53 is a novel N-terminal p53 isoform encoded by Δ133p53 transcript. FEBS Lett. 2010, 584, 4463–4468, doi:10.1016/j.febslet.2010.10.005. Available online: https://www.ncbi.nlm.nih.gov/pubmed/20937277 (accessed on 5 October 2020).
- Lowe, S.W.; E Ruley, H. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993, 7, 535–545, doi:10.1101/gad.7.4.535. Available online: https://www.ncbi.nlm.nih.gov/pubmed/8384579 (accessed on 5 October 2020).
- Yin, Y.; Stephen, C.W.; Luciani, M.G.; Fåhraeus, R. p53 stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat. Cell Biol. 2002, 4, 462–467, doi:10.1038/ncb801. Available online: https://www.ncbi.nlm.nih.gov/pubmed/12032546 (accessed on 5 October 2020).
- Bulavin, D.V.; Saito, S.; Hollander, M.C.; Sakaguchi, K.; Anderson, C.W.; Appella, E.; Jr, A.J.F. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 1999, 18, 6845–6854, doi:10.1093/emboj/18.23.6845. Available online: https://www.ncbi.nlm.nih.gov/pubmed/10581258 (accessed on 5 October 2020).
- D’Orazi, G.; Cecchinelli, B.; Bruno, T.; Manni, I.; Higashimoto, Y.; Saito, S.; Gostissa, M.; Coen, S.; Marchetti, A.; Del Sal, G.; et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 2002, 4, 11–19, doi:10.1038/ncb714. Available online: https://www.ncbi.nlm.nih.gov/pubmed/11780126 (accessed on 5 October 2020).
- Yoshida, K.; Liu, H.; Miki, Y. Protein Kinase C δ Regulates Ser46Phosphorylation of p53 Tumor Suppressor in the Apoptotic Response to DNA Damage. J. Biol. Chem. 2005, 281, 5734–5740, doi:10.1074/jbc.m512074200. Available online: https://www.ncbi.nlm.nih.gov/pubmed/16377624 (accessed on 5 October 2020).
- Taira, N.; Nihira, K.; Yamaguchi, T.; Miki, Y.; Yoshida, K. DYRK2 Is Targeted to the Nucleus and Controls p53 via Ser46 Phosphorylation in the Apoptotic Response to DNA Damage. Mol. Cell 2007, 25, 725–738, doi:10.1016/j.molcel.2007.02.007. Available online: https://www.ncbi.nlm.nih.gov/pubmed/17349958 (accessed on 5 October 2020).
- Graupner, V.; Schulze-Osthoff, K.; Essmann, F.; Jänicke, R.U. Functional characterization of p53? and p53?, two isoforms of the tumor suppressor p53. Cell Cycle 2009, 8, 1238–1248, doi:10.4161/cc.8.8.8251. Available online: https://www.ncbi.nlm.nih.gov/pubmed/19305147 (accessed on 5 October 2020).
- Brosh, R.; Rotter, V. When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer 2009, 9, 701–713, doi:10.1038/nrc2693. Available online: https://www.ncbi.nlm.nih.gov/pubmed/19693097 (accessed on 5 October 2020).
- Strano, S.; Dell’Orso, S.; Di Agostino, S.; Fontemaggi, G.; Sacchi, A.; Blandino, G. Mutant p53: an oncogenic transcription factor. Oncogene 2007, 26, 2212–2219, doi:10.1038/sj.onc.1210296. Available online: https://www.ncbi.nlm.nih.gov/pubmed/17401430 (accessed on 5 October 2020).
- Bargonetti, J.; Prives, C. Gain-of-function mutant p53: history and speculation. J. Mol. Cell Biol. 2019, 11, 605–609, doi:10.1093/jmcb/mjz067. Available online: https://www.ncbi.nlm.nih.gov/pubmed/31283823 (accessed on 5 October 2020).
- Chang, C.-J.; Chao, C.-H.; Xia, W.; Yang, J.-Y.; Xiong, Y.; Li, C.-W.; Yu, W.-H.; Rehman, S.K.; Hsu, J.L.; Lee, H.-H.; et al. p53 regulates epithelial–mesenchymal transition and stem cell properties through modulating miRNAs. Nat. Cell Biol. 2011, 13, 317–323, doi:10.1038/ncb2173. Available online: https://www.ncbi.nlm.nih.gov/pubmed/21336307 (accessed on 5 October 2020).
- Tamura, M.; Sasaki, Y.; Kobashi, K.; Takeda, K.; Nakagaki, T.; Idogawa, M.; Tokino, T. CRKL oncogene is downregulated by p53 through miR‐200s. Cancer Sci. 2015, 106, 1033–1040, doi:10.1111/cas.12713. Available online: https://www.ncbi.nlm.nih.gov/pubmed/26079153 (accessed on 5 October 2020).
- Alam, F.; Mezhal, F.; El Hasasna, H.; A Nair, V.; Aravind, S.R.; Ayad, M.S.; El-Serafi, A.; Abdel-Rahman, W.M. The role of p53-microRNA 200-Moesin axis in invasion and drug resistance of breast cancer cells. Tumor Biol. 2017, 39, doi:10.1177/1010428317714634. Available online: https://www.ncbi.nlm.nih.gov/pubmed/28933253 (accessed on 5 October 2020).
- Soond, S.M.; Kozhevnikova, M.V.; Frolova, A.S.; Savvateeva, L.V.; Plotnikov, E.Y.; Townsend, P.A.; Han, Y.-P.; Zamyatnin, A.A. Lost or Forgotten: The nuclear cathepsin protein isoforms in cancer. Cancer Lett. 2019, 462, 43–50, doi:10.1016/j.canlet.2019.07.020. Available online: https://www.ncbi.nlm.nih.gov/pubmed/31381961 (accessed on 5 October 2020).
- Gocheva, V.; Joyce, J.A. Cysteine cathepsins and the cutting edge of cancer invasion. Cell Cycle 2007, 6, 60–64, doi:10.4161/cc.6.1.3669. Available online: https://www.ncbi.nlm.nih.gov/pubmed/17245112 (accessed on 5 October 2020).
- E Koblinski, J.; Ahram, M.; Sloane, B.F. Unraveling the role of proteases in cancer. Clin. Chim. Acta 2000, 291, 113–135, doi:10.1016/s0009-8981(99)00224-7. Available online: https://www.ncbi.nlm.nih.gov/pubmed/10675719 (accessed on 5 October 2020).
- Cirman, T.; Orešić, K.; Mazovec, G.D.; Turk, V.; Reed, J.C.; Myers, R.M.; Salvesen, G.S.; Turk, B. Selective Disruption of Lysosomes in HeLa Cells Triggers Apoptosis Mediated by Cleavage of Bid by Multiple Papain-like Lysosomal Cathepsins. J. Biol. Chem. 2003, 279, 3578–3587, doi:10.1074/jbc.m308347200. Available online: https://www.ncbi.nlm.nih.gov/pubmed/14581476 (accessed on 5 October 2020).
- Duncan, E.M.; Muratore-Schroeder, T.L.; Cook, R.G.; Garcia, B.A.; Shabanowitz, J.; Hunt, D.F.; Allis, C.D. Cathepsin L Proteolytically Processes Histone H3 During Mouse Embryonic Stem Cell Differentiation. Cell 2008, 135, 284–294, doi:10.1016/j.cell.2008.09.055. Available online: https://www.ncbi.nlm.nih.gov/pubmed/18957203 (accessed on 5 October 2020).
- Adams-Cioaba, M.A.; Krupa, J.C.; Xu, C.; Mort, J.S.; Min, J. Structural basis for the recognition and cleavage of histone H3 by cathepsin L. Nat. Commun. 2011, 2, 197, doi:10.1038/ncomms1204. Available online: https://www.ncbi.nlm.nih.gov/pubmed/21326229 (accessed on 5 October 2020).
- Goulet, B.; Baruch, A.; Moon, N.-S.; Poirier, M.; Sansregret, L.L.; Erickson, A.; Bogyo, M.; Nepveu, A. A Cathepsin L Isoform that Is Devoid of a Signal Peptide Localizes to the Nucleus in S Phase and Processes the CDP/Cux Transcription Factor. Mol. Cell 2004, 14, 207–219, doi:10.1016/s1097-2765(04)00209-6. Available online: https://www.ncbi.nlm.nih.gov/pubmed/15099520 (accessed on 5 October 2020).
- Wang, L.; Zhao, Y.; Xiong, Y.; Wang, W.; Fei, Y.; Tan, C.; Liang, Z. K-ras mutation promotes ionizing radiation-induced invasion and migration of lung cancer in part via the Cathepsin L/CUX1 pathway. Exp. Cell Res. 2018, 362, 424–435, doi:10.1016/j.yexcr.2017.12.006. Available online: https://www.ncbi.nlm.nih.gov/pubmed/29246726 (accessed on 5 October 2020).
- Burton, L.J.; Dougan, J.; Jones, J.; Smith, B.N.; Randle, D.; Henderson, V.; A Odero-Marah, V. Targeting the Nuclear Cathepsin L CCAAT Displacement Protein/Cut Homeobox Transcription Factor-Epithelial Mesenchymal Transition Pathway in Prostate and Breast Cancer Cells with the Z-FY-CHO Inhibitor. Mol. Cell. Biol. 2016, 37, doi:10.1128/mcb.00297-16. Available online: https://www.ncbi.nlm.nih.gov/pubmed/27956696 (accessed on 5 October 2020).
- Wu, R.-W.; Lian, W.-S.; Chen, Y.-S.; Kuo, C.-W.; Ke, H.-C.; Hsieh, C.-K.; Wang, S.-Y.; Ko, J.-Y.; Wang, F.-S. MicroRNA-29a Counteracts Glucocorticoid Induction of Bone Loss through Repressing TNFSF13b Modulation of Osteoclastogenesis. Int. J. Mol. Sci. 2019, 20, 5141, doi:10.3390/ijms20205141. Available online: https://www.ncbi.nlm.nih.gov/pubmed/31627291 (accessed on 5 October 2020).
- Hu, Z.-Q.; Rao, C.-L.; Tang, M.-L.; Zhang, Y.; Lu, X.-X.; Chen, J.-G.; Mao, C.; Deng, L.; Li, Q.; Mao, X.-H. Rab32 GTPase, as a direct target of miR-30b/c, controls the intracellular survival of Burkholderia pseudomallei by regulating phagosome maturation. PLOS Pathog. 2019, 15, doi:10.1371/journal.ppat.1007879. Available online: https://www.ncbi.nlm.nih.gov/pubmed/31199852 (accessed on 5 October 2020).
- Huang, Y.; Ren, K.; Yao, T.; Zhu, H.; Xu, Y.; Ye, H.; Chen, Z.; Lv, J.; Shen, S.; Ma, J. MicroRNA-25-3p regulates osteoclasts through nuclear factor I X. Biochem. Biophys. Res. Commun. 2019, 522, 74–80, doi:10.1016/j.bbrc.2019.11.043. Available online: https://www.ncbi.nlm.nih.gov/pubmed/31740002 (accessed on 5 October 2020).
- Ho, K.-H.; Cheng, C.-H.; Chou, C.-M.; Chen, P.-H.; Liu, A.-J.; Lin, C.-W.; Shih, C.-M.; Chen, K.-C. miR-140 targeting CTSB signaling suppresses the mesenchymal transition and enhances temozolomide cytotoxicity in glioblastoma multiforme. Pharmacol. Res. 2019, 147, doi:10.1016/j.phrs.2019.104390. Available online: https://www.ncbi.nlm.nih.gov/pubmed/31398406 (accessed on 5 October 2020).
- Li, K.; Chen, S.; Cai, P.; Chen, K.; Li, L.; Yang, X.; Yi, J.; Luo, X.; Du, Y.; Zheng, H. MiRNA-483–5p is involved in the pathogenesis of osteoporosis by promoting osteoclast differentiation. Mol. Cell. Probes 2020, 49, doi:10.1016/j.mcp.2019.101479. Available online: https://www.ncbi.nlm.nih.gov/pubmed/31706013 (accessed on 5 October 2020).
- Dinesh, P.; Kalaiselvan, S.; Sujitha, S.; Rasool, M. miR‐506‐3p alleviates uncontrolled osteoclastogenesis via repression of RANKL/NFATc1 signaling pathway. J. Cell. Physiol. 2020, 235, 9497–9509, doi:10.1002/jcp.29757. Available online: https://www.ncbi.nlm.nih.gov/pubmed/32372426 (accessed on 5 October 2020).
- Zhao, Y.-F.; Han, M.-L.; Xiong, Y.-J.; Wang, L.; Fei, Y.; Shen, X.; Zhu, Y.; Liang, Z.-Q. A miRNA-200c/cathepsin L feedback loop determines paclitaxel resistance in human lung cancer A549 cells in vitro through regulating epithelial–mesenchymal transition. Acta Pharmacol. Sin. 2017, 39, 1034–1047, doi:10.1038/aps.2017.164. Available online: https://www.ncbi.nlm.nih.gov/pubmed/29219949 (accessed on 5 October 2020).
- Okamura, S.; Arakawa, H.; Tanaka, T.; Nakanishi, H.; Ng, C.C.; Taya, Y.; Monden, M.; Nakamura, Y. p53DINP1, a p53-Inducible Gene, Regulates p53-Dependent Apoptosis. Mol. Cell 2001, 8, 85–94, doi:10.1016/s1097-2765(01)00284-2. Available online: https://www.ncbi.nlm.nih.gov/pubmed/11511362 (accessed on 5 October 2020).
- Lu, H.-J.; Yan, J.; Jin, P.-Y.; Zheng, G.-H.; Qin, S.-M.; Wu, D.; Lu, J.; Zheng, Y.-L. MicroRNA-152 inhibits tumor cell growth while inducing apoptosis via the transcriptional repression of cathepsin L in gastrointestinal stromal tumor. Cancer Biomarkers 2018, 21, 711–722, doi:10.3233/cbm-170809. Available online: https://www.ncbi.nlm.nih.gov/pubmed/29278883 (accessed on 5 October 2020).
- Kozlowski, L.; Wojtukiewicz, M.Z.; Ostrowska, H. Cathepsin A activity in primary and metastatic human melanocytic tumors. Arch. Dermatol. Res. 2000, 292, 68–71, doi:10.1007/s004030050012. Available online: https://www.ncbi.nlm.nih.gov/pubmed/10749558 (accessed on 5 October 2020).
- Ni, S.; Weng, W.; Xu, M.; Wang, Q.; Tan, C.; Sun, H.; Wang, L.; Huang, D.; Du, X.; Sheng, W. miR-106b-5p inhibits the invasion and metastasis of colorectal cancer by targeting CTSA. OncoTargets Ther. 2018, 11, 3835–3845, doi:10.2147/ott.s172887. Available online: https://www.ncbi.nlm.nih.gov/pubmed/30013364 (accessed on 5 October 2020).
- Wang, Y.; Weng, T.; Gou, D.; Chen, Z.; Chintagari, N.R.; Liu, L. Identification of rat lung-specific microRNAs by microRNA microarray: valuable discoveries for the facilitation of lung research. BMC Genom. 2007, 8, 1–14, doi:10.1186/1471-2164-8-29. Available online: https://www.ncbi.nlm.nih.gov/pubmed/17250765 (accessed on 5 October 2020).
- Ceppi, P.; Mudduluru, G.; Kumarswamy, R.; Rapa, I.; Scagliotti, G.V.; Papotti, M.; Allgayer, H. Loss of miR-200c Expression Induces an Aggressive, Invasive, and Chemoresistant Phenotype in Non-Small Cell Lung Cancer. Mol. Cancer Res. 2010, 8, 1207–1216, doi:10.1158/1541-7786.mcr-10-0052. Available online: https://www.ncbi.nlm.nih.gov/pubmed/20696752 (accessed on 5 October 2020).
- Cittelly, D.M.; Dimitrova, I.; Howe, E.N.; Cochrane, D.R.; Jean, A.; Spoelstra, N.S.; Post, M.D.; Lu, X.; Broaddus, R.R.; Spillman, M.A.; et al. Restoration of miR-200c to Ovarian Cancer Reduces Tumor Burden and Increases Sensitivity to Paclitaxel. Mol. Cancer Ther. 2012, 11, 2556–2565, doi:10.1158/1535-7163.mct-12-0463. Available online: https://www.ncbi.nlm.nih.gov/pubmed/23074172 (accessed on 5 October 2020).
- Shi, L.; Zhang, S.; Wu, H.; Zhang, L.; Dai, X.; Hu, J.; Xue, J.; Liu, T.; Liang, Y.; Wu, G. MiR-200c Increases the Radiosensitivity of Non-Small-Cell Lung Cancer Cell Line A549 by Targeting VEGF-VEGFR2 Pathway. PLOS ONE 2013, 8, e78344, doi:10.1371/journal.pone.0078344. Available online: https://www.ncbi.nlm.nih.gov/pubmed/24205206 (accessed on 5 October 2020).
- Cortez, M.A.; Valdecanas, D.; Zhang, X.; Zhan, Y.; Bhardwaj, V.; A Calin, G.; Komaki, R.; Giri, D.K.; Quini, C.C.; Wolfe, T.; et al. Therapeutic Delivery of miR-200c Enhances Radiosensitivity in Lung Cancer. Mol. Ther. 2014, 22, 1494–1503, doi:10.1038/mt.2014.79. Available online: https://www.ncbi.nlm.nih.gov/pubmed/24791940 (accessed on 5 October 2020).
- Kopp, F.; Wagner, E.; Roidl, A. The proto-oncogene KRAS is targeted by miR-200c. Oncotarget 2013, 5, 185–195, doi:10.18632/oncotarget.1427. Available online: https://www.ncbi.nlm.nih.gov/pubmed/24368337 (accessed on 5 October 2020).
- Li, J.; Tan, Q.; Yan, M.; Liu, L.; Lin, H.; Zhao, F.; Bao, G.; Kong, H.; Ge, C.; Zhang, F.; et al. miRNA-200c inhibits invasion and metastasis of human non-small cell lung cancer by directly targeting ubiquitin specific peptidase 25. Mol. Cancer 2014, 13, 166, doi:10.1186/1476-4598-13-166. Available online: https://www.ncbi.nlm.nih.gov/pubmed/24997798 (accessed on 5 October 2020).
- Zhou, G.; Zhang, F.; Guo, Y.; Huang, J.; Xie, Y.; Yue, S.; Chen, M.; Jiang, H.; Li, M. miR-200c enhances sensitivity of drug-resistant non-small cell lung cancer to gefitinib by suppression of PI3K/Akt signaling pathway and inhibites cell migration via targeting ZEB1. Biomed. Pharmacother. 2017, 85, 113–119, doi:10.1016/j.biopha.2016.11.100. Available online: https://www.ncbi.nlm.nih.gov/pubmed/27930974 (accessed on 5 October 2020).
- Jiao, A.; Sui, M.; Zhang, L.; Sun, P.; Geng, D.; Zhang, W.; Wang, X.; Li, J. MicroRNA-200c inhibits the metastasis of non-small cell lung cancer cells by targeting ZEB2, an epithelial-mesenchymal transition regulator. Mol. Med. Rep. 2016, 13, 3349–3355, doi:10.3892/mmr.2016.4901. Available online: https://www.ncbi.nlm.nih.gov/pubmed/26935975 (accessed on 5 October 2020).
- Tejero, R.; Navarro, A.; Campayo, M.; Viñolas, N.; Marrades, R.M.; Cordeiro, A.; Ruíz-Martínez, M.; Santasusagna, S.; Molins, L.; Ramirez, J.; et al. miR-141 and miR-200c as Markers of Overall Survival in Early Stage Non-Small Cell Lung Cancer Adenocarcinoma. PLOS ONE 2014, 9, e101899, doi:10.1371/journal.pone.0101899. Available online: https://www.ncbi.nlm.nih.gov/pubmed/25003366 (accessed on 5 October 2020).
- Bai, T.; Dong, D.-S.; Pei, L. Synergistic antitumor activity of resveratrol and miR-200c in human lung cancer. Oncol. Rep. 2014, 31, 2293–2297, doi:10.3892/or.2014.3090. Available online: https://www.ncbi.nlm.nih.gov/pubmed/24647918 (accessed on 5 October 2020).
- Kim, M.K.; Jung, S.B.; Kim, J.-S.; Roh, M.S.; Lee, J.H.; Lee, E.H.; Lee, H.W. Expression of microRNA miR-126 and miR-200c is associated with prognosis in patients with non-small cell lung cancer. Virchows Archiv. 2014, 465, 463–471, doi:10.1007/s00428-014-1640-4. Available online: https://www.ncbi.nlm.nih.gov/pubmed/25124149 (accessed on 5 October 2020).
- Shao, Y.; Geng, Y.; Gu, W.; Huang, J.; Pei, H.; Jiang, J. Prognostic Role of Tissue and Circulating MicroRNA-200c in Malignant Tumors: A Systematic Review and Meta-Analysis. Cell. Physiol. Biochem. 2015, 35, 1188–1200, doi:10.1159/000373943. Available online: https://www.ncbi.nlm.nih.gov/pubmed/25766530 (accessed on 5 October 2020).
- Teng, Y.; Su, X.; Zhang, X.; Zhang, Y.; Li, C.; Niu, W.; Liu, C.; Qu, K. miRNA-200a/c as potential biomarker in epithelial ovarian cancer (EOC): evidence based on miRNA meta-signature and clinical investigations. Oncotarget 2016, 7, 81621–81633, doi:10.18632/oncotarget.13154. Available online: https://www.ncbi.nlm.nih.gov/pubmed/27835595 (accessed on 5 October 2020).
- Si, L.; Tian, H.; Yue, W.; Li, L.; Li, S.; Gao, C.; Qi, L. Potential use of microRNA-200c as a prognostic marker in non-small cell lung cancer. Oncol. Lett. 2017, 14, 4325–4330, doi:10.3892/ol.2017.6667. Available online: https://www.ncbi.nlm.nih.gov/pubmed/28943946 (accessed on 5 October 2020).
- Li, Y.; Huang, H.-C.; Chen, L.-Q.; Xu, L.-Y.; Li, E.-M.; Zhang, J.-J. Predictive biomarkers for response of esophageal cancer to chemo(radio)therapy: A systematic review and meta-analysis. Surg. Oncol. 2017, 26, 460–472, doi:10.1016/j.suronc.2017.09.003. Available online: https://www.ncbi.nlm.nih.gov/pubmed/29113666 (accessed on 5 October 2020).
- Zheng, Q.; Chen, C.; Guan, H.; Kang, W.; Yu, C. Prognostic role of microRNAs in human gastrointestinal cancer: A systematic review and meta-analysis. Oncotarget 2017, 8, 46611–46623, doi:10.18632/oncotarget.16679. Available online: https://www.ncbi.nlm.nih.gov/pubmed/28402940 (accessed on 5 October 2020).
- Shi, M.; Mu, Y.; Zhang, H.; Liu, M.; Wan, J.; Qin, X.; Li, C. MicroRNA-200 and microRNA-30 family as prognostic molecular signatures in ovarian cancer. Medicine 2018, 97, doi:10.1097/md.0000000000011505. Available online: https://www.ncbi.nlm.nih.gov/pubmed/30095616 (accessed on 5 October 2020).
- Chen, Y.; Song, Y.; Wang, Z.-N.; Yue, Z.; Xu, H.-M.; Xing, C.; Liu, Z. Altered Expression of MiR-148a and MiR-152 in Gastrointestinal Cancers and Its Clinical Significance. J. Gastrointest. Surg. 2010, 14, 1170–1179, doi:10.1007/s11605-010-1202-2. Available online: https://www.ncbi.nlm.nih.gov/pubmed/20422307 (accessed on 5 October 2020).
- Ge, S.; Wang, D.; Kong, Q.; Gao, W.; Sun, J. Function of miR-152 as a Tumor Suppressor in Human Breast Cancer by Targeting PIK3CA. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 1363–1371, doi:10.3727/096504017x14878536973557. Available online: https://www.ncbi.nlm.nih.gov/pubmed/28247844 (accessed on 5 October 2020).
- Li, B.; Xie, Z.; Li, B. miR-152 functions as a tumor suppressor in colorectal cancer by targeting PIK3R3. Tumor Biol. 2016, 37, 10075–10084, doi:10.1007/s13277-016-4888-2. Available online: https://www.ncbi.nlm.nih.gov/pubmed/26820128 (accessed on 5 October 2020).
- Song, Y.; Zhang, M.; Lu, M.M.; Qu, L.Y.; Xu, S.G.; Li, Y.Z.; Wang, M.Y.; Zhu, H.F.; Zhang, Z.Y.; He, G.Y.; et al. EPAS1 targeting by miR-152-3p in Paclitaxel-resistant Breast Cancer. J. Cancer 2020, 11, 5822–5830, doi:10.7150/jca.46898. Available online: https://www.ncbi.nlm.nih.gov/pubmed/32913475 (accessed on 5 October 2020).
- Zhai, R.; Kan, X.; Wang, B.; Du, H.; Long, Y.; Wu, H.; Tao, K.; Wang, G.; Bao, L.; Li, F.; et al. miR-152 suppresses gastric cancer cell proliferation and motility by targeting CD151. Tumor Biol. 2014, 35, 11367–11373, doi:10.1007/s13277-014-2471-2. Available online: https://www.ncbi.nlm.nih.gov/pubmed/25119599 (accessed on 5 October 2020).
- Xu, Q.; Jiang, Y.; Yin, Y.; Li, Q.; He, J.; Jing, Y.; Qi, Y.-T.; Xu, Q.; Li, W.; Lu, B.; et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J. Mol. Cell Biol. 2012, 5, 3–13, doi:10.1093/jmcb/mjs049. Available online: https://www.ncbi.nlm.nih.gov/pubmed/22935141 (accessed on 5 October 2020).
- Wang, Y.; Wang, D.; Xie, G.; Yin, Y.; Zhao, E.; Tao, K.; Li, R. MicroRNA-152 regulates immune response via targeting B7-H1 in gastric carcinoma. Oncotarget 2017, 8, 28125–28134, doi:10.18632/oncotarget.15924. Available online: https://www.ncbi.nlm.nih.gov/pubmed/28427226 (accessed on 5 October 2020).
- Yin, T.; Liu, M.-M.; Jin, R.-T.; Kong, J.; Wang, S.-H.; Sun, W.-B. miR-152-3p Modulates hepatic carcinogenesis by targeting cyclin-dependent kinase 8. Pathol. Res. Pract. 2019, 215, 152406, doi:10.1016/j.prp.2019.03.034. Available online: https://www.ncbi.nlm.nih.gov/pubmed/30967300 (accessed on 5 October 2020).
- Wang, L.; Wang, Y.; Lin, J. MiR-152-3p promotes the development of chronic myeloid leukemia by inhibiting p27. Eur. Rev. Med Pharmacol. Sci. 2018, 22, 8789–8796. Available online: https://www.ncbi.nlm.nih.gov/pubmed/30575920 (accessed on 5 October 2020).
- Wang, M.; Wu, Q.; Fang, M.; Huang, W.; Zhu, H. miR-152-3p Sensitizes Glioblastoma Cells Towards Cisplatin Via Regulation of SOS1. OncoTargets Ther. 2019, 12, 9513–9525, doi:10.2147/OTT.S210732. Available online: https://www.ncbi.nlm.nih.gov/pubmed/31807027 (accessed on 5 October 2020).
- Zhu, X.; Shen, Z.; Man, D.; Ruan, H.; Huang, S. miR-152-3p Affects the Progression of Colon Cancer via the KLF4/IFITM3 Axis. Comput. Math. Methods Med. 2020, 2020, 1–10, doi:10.1155/2020/8209504. Available online: https://www.ncbi.nlm.nih.gov/pubmed/32952601 (accessed on 5 October 2020).
- Sanfiorenzo, C.; Ilie, M.; Belaid, A.; Barlesi, F.; Mouroux, J.; Marquette, C.-H.; Brest, P.; Hofman, P. Two Panels of Plasma MicroRNAs as Non-Invasive Biomarkers for Prediction of Recurrence in Resectable NSCLC. PLOS ONE 2013, 8, doi:10.1371/journal.pone.0054596. Available online: https://www.ncbi.nlm.nih.gov/pubmed/23342174 (accessed on 5 October 2020).
- You, W.; Zhang, X.; Ji, M.; Yu, Y.; Chen, C.; Xiong, Y.; Liu, Y.; Sun, Y.; Tan, C.; Zhang, H.; et al. MiR-152-5p as a microRNA passenger strand special functions in human gastric cancer cells. Int. J. Biol. Sci. 2018, 14, 644–653, doi:10.7150/ijbs.25272. Available online: https://www.ncbi.nlm.nih.gov/pubmed/29904279 (accessed on 5 October 2020).
- Matin, F.; BioResource, A.P.C.; Jeet, V.; Moya, L.; Selth, L.A.; Chambers, S.; Clements, J.A.; Batra, J. A Plasma Biomarker Panel of Four MicroRNAs for the Diagnosis of Prostate Cancer. Sci. Rep. 2018, 8, 1–15, doi:10.1038/s41598-018-24424-w. Available online: https://www.ncbi.nlm.nih.gov/pubmed/29703916 (accessed on 5 October 2020).
- Chen, H.; Liu, H.; Zou, H.; Chen, R.; Dou, Y.; Sheng, S.; Dai, S.; Ai, J.; Melson, J.; Kittles, R.A.; et al. Evaluation of Plasma miR-21 and miR-152 as Diagnostic Biomarkers for Common Types of Human Cancers. J. Cancer 2016, 7, 490–499, doi:10.7150/jca.12351. Available online: https://www.ncbi.nlm.nih.gov/pubmed/26958084 (accessed on 5 October 2020).
- Li, X.; Zou, W.; Wang, Y.; Liao, Z.; Li, L.; Zhai, Y.; Zhang, L.; Gu, S.; Zhao, X.H. Plasma‐based microRNA signatures in early diagnosis of breast cancer. Mol. Genet. Genom. Med. 2020, 8, doi:10.1002/mgg3.1092. Available online: https://www.ncbi.nlm.nih.gov/pubmed/32124558 (accessed on 5 October 2020).
- Cai, K.; Wang, Y.; Bao, X. MiR-106b promotes cell proliferation via targeting RB in laryngeal carcinoma. J. Exp. Clin. Cancer Res. 2011, 30, 73, doi:10.1186/1756-9966-30-73. Available online: https://www.ncbi.nlm.nih.gov/pubmed/21819631 (accessed on 5 October 2020).
- Zhai, Z.; Wu, F.; Chuang, A.Y.; Kwon, J.H. miR-106b Fine Tunes ATG16L1 Expression and Autophagic Activity in Intestinal Epithelial HCT116 Cells. Inflamm. Bowel Dis. 2013, 19, 2295–2301, doi:10.1097/mib.0b013e31829e71cf. Available online: https://www.ncbi.nlm.nih.gov/pubmed/23899543 (accessed on 5 October 2020).
- Lu, C.; Chen, J.; Xu, H.; Zhou, X.; He, Q.; Li, Y.; Jiang, G.; Shan, Y.; Xue, B.; Zhao, R.; et al. MIR106B and MIR93 Prevent Removal of Bacteria from Epithelial Cells by Disrupting ATG16L1-Mediated Autophagy. Gastroenterology 2014, 146, 188–199, doi:10.1053/j.gastro.2013.09.006. Available online: https://www.ncbi.nlm.nih.gov/pubmed/24036151 (accessed on 5 October 2020).
- Guo, J.; Miao, Y.; Xiao, B.; Huan, R.; Jiang, Z.; Meng, D.; Wang, Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J. Gastroenterol. Hepatol. 2009, 24, 652–657, doi:10.1111/j.1440-1746.2008.05666.x. Available online: https://www.ncbi.nlm.nih.gov/pubmed/19175831 (accessed on 5 October 2020).
- Zheng, L.; Zhang, Y.; Liu, Y.; Zhou, M.; Lu, Y.; Yuan, L.; Zhang, C.; Hong, M.; Wang, S.; Li, X. MiR-106b induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer. J. Transl. Med. 2015, 13, 1–13, doi:10.1186/s12967-015-0592-z. Available online: https://www.ncbi.nlm.nih.gov/pubmed/26238857 (accessed on 5 October 2020).
- Liu, H.; Liu, Y.; Sun, P.; Leng, K.; Xu, Y.; Mei, L.; Han, P.; Zhang, B.; Yao, K.; Li, C.; et al. Colorectal cancer-derived exosomal miR-106b-3p promotes metastasis by down-regulating DLC-1 expression. Clin. Sci. 2020, 134, 419–434, doi:10.1042/cs20191087. Available online: https://www.ncbi.nlm.nih.gov/pubmed/32065214 (accessed on 5 October 2020).
- Pan, M.; Chen, Q.; Lu, Y.; Wei, F.; Chen, C.; Tang, G.; Huang, H. MiR-106b-5p Regulates the Migration and Invasion of Colorectal Cancer Cells by Targeting FAT4. Biosci. Rep. 2020, 40, doi:10.1042/bsr20200098. Available online: https://www.ncbi.nlm.nih.gov/pubmed/33063118 (accessed on 5 October 2020).
- Wang, Y.X.; Zhang, X.Y.; Zhang, B.F.; Yang, C.Q.; Chen, X.M.; Gao, H.J. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J. Dig. Dis. 2010, 11, 50–54, doi:10.1111/j.1751-2980.2009.00413.x. Available online: https://www.ncbi.nlm.nih.gov/pubmed/20132431 (accessed on 5 October 2020).
- Nishida, N.; Nagahara, M.; Sato, T.; Mimori, K.; Sudo, T.; Tanaka, F.; Shibata, K.; Ishii, H.; Sugihara, K.; Doki, Y.; et al. Microarray Analysis of Colorectal Cancer Stromal Tissue Reveals Upregulation of Two Oncogenic miRNA Clusters. Clin. Cancer Res. 2012, 18, 3054–3070, doi:10.1158/1078-0432.ccr-11-1078. Available online: https://www.ncbi.nlm.nih.gov/pubmed/22452939 (accessed on 5 October 2020).
- Wang, Y.-X.; Lang, F.; Liu, Y.-X.; Yang, C.-Q.; Gao, H. In situ hybridization analysis of the expression of miR-106b in colonic cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 786–792. Available online: https://www.ncbi.nlm.nih.gov/pubmed/25755775 (accessed on 5 October 2020).
- Zhang, G.-J.; Li, J.-S.; Zhou, H.; Xiao, H.-X.; Li, Y.; Zhou, T. MicroRNA-106b promotes colorectal cancer cell migration and invasion by directly targeting DLC1. J. Exp. Clin. Cancer Res. 2015, 34, 1–11, doi:10.1186/s13046-015-0189-7. Available online: https://www.ncbi.nlm.nih.gov/pubmed/26223867 (accessed on 5 October 2020).
- Gu, L.; Li, H.; Chen, L.; Ma, X.; Gao, Y.; Li, X.; Zhang, Y.; Fan, Y.; Zhang, X. MicroRNAs as prognostic molecular signatures in renal cell carcinoma: a systematic review and meta-analysis. Oncotarget 2015, 6, 32545–32560, doi:10.18632/oncotarget.5324. Available online: https://www.ncbi.nlm.nih.gov/pubmed/26416448 (accessed on 5 October 2020).
- Liu, D.; Wan, X.; Shan, X.; Fan, R.; Zha, W. Drugging the “undruggable” microRNAs. Cell. Mol. Life Sci. 2020, 1–11, doi:10.1007/s00018-020-03676-8. Available online: https://www.ncbi.nlm.nih.gov/pubmed/33052435 (accessed on 5 October 2020).
