Assembly theory is a framework for quantifying evolution and complexity. It, therefore, spans various scientific disciplines, including physics, chemistry, biology, and information theory. Assembly theory is rooted in the assembly of an object from a set of basic building units, forming an initial assembly pool and from subunits that entered the assembly pool in previous assembly steps. Hence, the object is defined not as a set of point particles but by the history of its assembly, where the assembly index is the smallest number of steps required to assemble the object.
- assembly theory
- complexity
- origin of life
- emergent dimensionality
- mathematical physics
Assembly theory was formulated in 2017[1], introducing the concept of assembly index (initially called "pathway complexity") of an object as the smallest number of steps required to assemble this object from a set of basic building units, forming an initial assembly pool, and from subunits that entered the assembly pool in previous assembly steps. The assembly index is, therefore, a measure of the complexity of the object, which is computable[2], unlike Kolmogorov complexity, for example, and captures the structural information about the object, unlike Shannon entropy. The theoretical background for the theory was researched[2] based on directed multigraphs, showing that the assembly index of an object is computable for all finite objects. In particular, determining the assembly index is NP-complete[3].
Consider two binary strings C = [01010101] and D = [00010111] and the initial assembly pool containing two bits 0 and 1. Both strings have the same length N = 8 and the same Shannon entropy H(C) = H(D) = log2(2) = 1. However, the assembly index of the first string is a(C) = 3 (In step 1, assemble "01" and put it into the assembly pool, in step 2 assemble "01" with "01" taken from the assembly pool and put "0101" into the assembly pool, and in step 3 assemble "0101" assembled in the second step with "0101" taken from the assembly pool), while the assembly index of the second string is a(D) = 6, since only the substring "01" can be reused from the assembly pool[4].
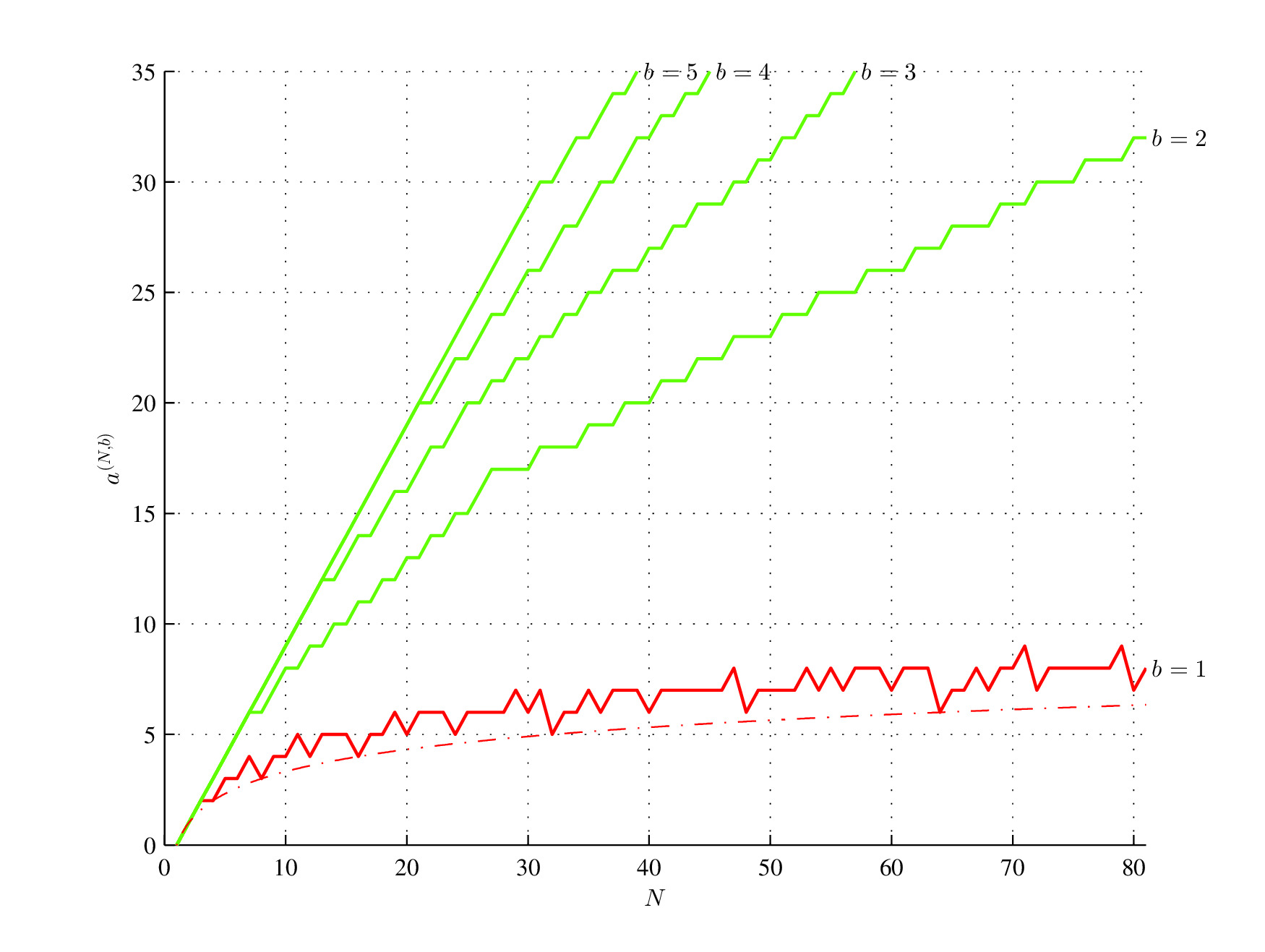
Minimum (red; log2(N), red, dash-dot) and maximum (green) assembly index for 1 ≤ b ≤ 5 and 0 < N ≤ 81. N is the string length; b is the alphabet size.
Basic building units depend on a particular application of the assembly theory. In chemistry, it has found applications in drug discovery[5]. Furthermore, the theoretical value of the assembly index of a molecule, where the initial assembly pool contains chemical bonds, can be experimentally confirmed using tandem mass spectrometry, nuclear magnetic resonance, or infrared spectroscopy[6][7]. Therefore, the assembly index is the universal threshold between abiotic and biotic molecules and a robust, simple biosignature for distinguishing random, abiotic objects from biologically or technologically assembled ones, as only biotic samples can have an assembly index above 15. The more complex a given object, the less likely an identical copy can exist without some information-driven mechanism that generates that object[8].
References
- Stuart M. Marshall; Alastair R. G. Murray; Leroy Cronin; A probabilistic framework for identifying biosignatures using Pathway Complexity. Philos. Trans. R. Soc. A: Math. Phys. Eng. Sci. 2017, 375, 20160342, .
- Stuart M. Marshall; Douglas G. Moore; Alastair R. G. Murray; Sara I. Walker; Leroy Cronin; Formalising the Pathways to Life Using Assembly Spaces. Entropy 2022, 24, 884, .
- Masierak, Piotr; Computational Complexity of Determining theAssembly Index. IPI Letters 2026, 4, 9-12, .
- Szymon Łukaszyk; Wawrzyniec Bieniawski; Assembly Theory of Binary Messages. Math. 2024, 12, 1600, .
- Yu Liu; Cole Mathis; Michał Dariusz Bajczyk; Stuart M. Marshall; Liam Wilbraham; Leroy Cronin; Exploring and mapping chemical space with molecular assembly trees. Sci. Adv. 2021, 7, eabj2465, .
- Stuart M. Marshall; Cole Mathis; Emma Carrick; Graham Keenan; Geoffrey J. T. Cooper; Heather Graham; Matthew Craven; Piotr S. Gromski; Douglas G. Moore; Sara. I. Walker; et al. Identifying molecules as biosignatures with assembly theory and mass spectrometry. Nat. Commun. 2021, 12, 1-9, .
- Michael Jirasek; Abhishek Sharma; Jessica R. Bame; S. Hessam M. Mehr; Nicola Bell; Stuart M. Marshall; Cole Mathis; Alasdair MacLeod; Geoffrey J. T. Cooper; Marcel Swart; et al. Investigating and Quantifying Molecular Complexity Using Assembly Theory and Spectroscopy. ACS Central Sci. 2024, 10, 1054-1064, .
- Abhishek Sharma; Dániel Czégel; Michael Lachmann; Christopher P. Kempes; Sara I. Walker; Leroy Cronin; Assembly theory explains and quantifies selection and evolution. Nat. 2023, 622, 321-328, .
- Masierak, Piotr; Computational Complexity of Determining theAssembly Index. IPI Letters 2026, 4, 9-12, .
