Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biochemistry & Molecular Biology
Breast cancer is a complex and enigmatic disease caused by a series of alterations in genes that control cell growth and proliferation. miRNA was discovered by Ambros and co-workers in Caenorhabditis elegans (Nematode), during their genetic study to investigate defects in the temporal control of C. elegans development.
- breast cancer
- microRNA
- biomarkers
1. Introduction
Breast cancer is a disease caused by genetic aberrations and deleterious environmental exposure [1]. Breast cancer is a complex and enigmatic disease caused by a series of alterations in genes that control cell growth and proliferation. Breast cancer is the most common cancer in women and is a heterogeneous disease with diverse molecular subtypes and clinical presentations. Despite advances in treatment, it is the leading cause of cancer mortality in women worldwide. Globally, it accounts for 16 percent of cancer deaths, and has over 20 distinct subtypes that differ genetically, morphologically, and clinically [2,3]. About 90% of these deaths are due to metastases. Currently, breast cancer is the fourth leading cause of cancer-related death, with 2.3 million new cases in 2022 worldwide [4]. The metastatic spread of breast cancer, typically to the bone, lung, liver, and brain, accounts for most cancer-related deaths [5]. There are few treatment strategies for metastatic breast cancer, which is incurable with a median survival rate of 2–3 years [6]. Breast cancer cases are increasing alarmingly, underscoring the importance of treating the disease in many ways. Several trials have been conducted for the successful development of drugs for breast cancers over the past few decades. As a result of inadequate early detection, breast cancer patients have a high mortality rate and recurrence rate. Thus, to improve disease outcomes and prolong patient survival, it is vital to find novel early prognostic and diagnostic biomarkers and effective therapeutic methods [7].
Recent research studies have shed light on the progression of breast cancer, its pathogenesis, and the molecular pathways involved in proliferation. As evidenced by recent research, molecular marker-based targeted therapies using miRNAs may improve the prognosis and diagnosis of a wide range of diseases, including breast cancer [8]. A growing body of evidence suggests that miRNAs play a critical role in tumorigenesis and breast cancer development. These molecules are altered in different tumorigenic processes of breast cancer. miRNAs are small, noncoding RNA molecules that regulate gene expression through interfering with transcription. miRNAs play a significant role in regulating a variety of cellular processes, such as angiogenesis, apoptosis, and the cell cycle. Due to their ability to modulate multiple targets within these pathways, they play a significant role in maintaining cellular homeostasis. It is often observed that dysregulation of miRNAs in these processes contributes to various diseases, including breast cancer, which makes them potential therapeutic targets or diagnostic markers for cancers [9]. It is important to understand the complex network of miRNA-mediated regulation to gain insight into the molecular mechanisms that govern these vital processes within cells.
miRNA was discovered by Ambros and co-workers in Caenorhabditis elegans (Nematode), during their genetic study to investigate defects in the temporal control of C. elegans development [10]. Recent studies have demonstrated that miRNAs play critical roles in the development of breast cancer, differentiation, proliferation, and other physiological processes [11]. A growing body of evidence suggests that miRNAs might have very important clinical implications.
2. miRNA Biogenesis and Maturation
Pri-miRNAs are processed step-by-step in the cytoplasm and nucleus during the biogenesis of miRNAs. In humans, miRNA gene transcription takes place within the nucleus following the cleavage of the ~80 nucleotide stem-loop pre-microRNA precursor performed by the microprocessor complex consisting of Drosha, an RNase III-type nuclease, a double strand RNA-binding protein co-factor, and the DiGeorge syndrome critical region 8 gene (DGCR8) (Figure 1). The complex cleaves the pri-miRNA to generate a shorter hairpin-shaped precursor called precursor miRNA (pre-miRNA). The pri-miRNAs are processed into 60–70 nucleotide hairpin structures (pre-miRNAs) and are exported from the nucleus to the cytoplasm, supported by the nucleocytoplasmic shuttle protein Exportin-5 in a Ran-GTP-dependent manner. The enzyme Dicer further breaks down the pre-miRNA into a double-stranded RNA duplex in the cytoplasm. The mature miRNA is one of the two strands of the duplex that is chosen and loaded into the RNA-induced silencing complex (RISC) [17,18].
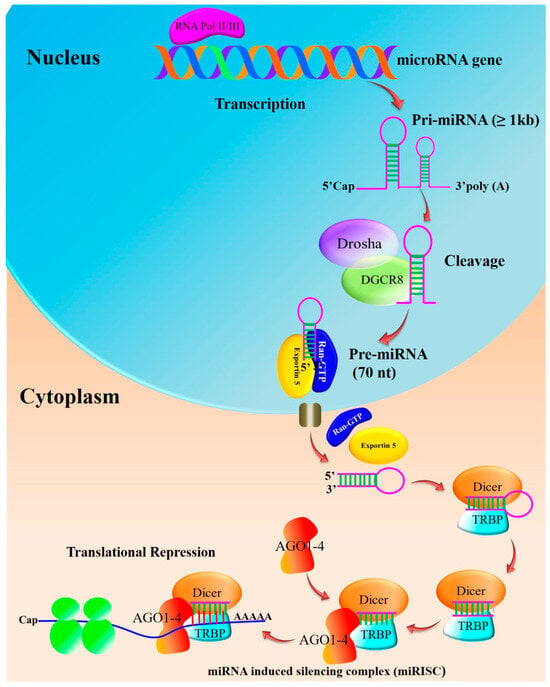
Figure 1. Biosynthesis of miRNA: Pre-miRNAs are further cleaved into an asymmetric duplex using the action of Dicer and accessory proteins. Transactivation-responsive RNA-binding protein (TRBP) and PACT in humans remove the loop sequence by forming a short-lived asymmetric duplex intermediate (miRNA: miRNA), with a duplex of about 22 nucleotides in length. This pre-cursor is cleaved to generate ~21–25-nucleotide mature miRNAs. The mature miRNA is loaded into the miRNA-induced silencing complex (miRISC), which binds to target mRNA, resulting in either the degradation of mRNA or the blockage of translation without mRNA degradation (adapted from [17,18]).
By base-pairing with complementary sequences in the 3’ untranslated region (UTR) of the target mRNA, the mature miRNA directs the RISC complex to its target mRNAs. As a result of this interaction, the target gene may be downregulated due to translational repression or mRNA degradation. Multiple steps and molecular players are involved in miRNA biogenesis [17]. Breast cancer has been associated with dysregulation of miRNA biogenesis or its function.
3. miRNA Targets and Their Role in Breast Cancer
Discovering the regulatory network regulated by miRNAs requires identification of miRNA-mRNA target interactions. The development of miRNA target prediction algorithms is based on several approaches. Statistical inference based on machine learning and algorithms derived from characteristics of the mRNA sequence and/or based on the miRNA-mRNA interaction are the two major categories. Pairings of miRNA and mRNA seed sequences can be analyzed and evaluated. Instead of making “de novo” predictions based on sequence features, the aim of machine learning is to classify miRNA targets that reference miRNA-mRNA duplexes with known biological significance [19]. miRNA sequences are complementary to 3′-UTR sequences of mRNA targets. For miRNA to bind to target mRNAs, the seed sequence of the miRNA 5′ region is essential. Specific seed region characteristics, as well as those in proximity, have been linked to specific effects on miRNA-induced gene repression [19].
Watson–Crick Pairing between miRNA and mRNA is needed for most target prediction algorithms. miRNA prediction tools include miRNAFinder, miRscan, miRbase, miRTarBase, and SSC profiler [20,21]. Most of them are focused on miRNA conservation characteristics across ecosystems. The prediction of miRNA in a wide range of species from the animal and plant kingdoms have proven successful with this technique.
The miRNAs play an important role in the regulation of gene expression, and the dysregulation of these molecules has been implicated in various stages of breast cancer development. Breast cancer miRNAs are classified based on their expression patterns, functional roles, and clinical implications [22,23]. miRNAs can be classified into two main categories established on their effects on tumorigenesis:
-
Oncogenic miRNAs (OncomiRs): these miRNAs are often upregulated in breast cancer and promote tumorigenesis by inhibiting tumor-suppressor genes or regulatory pathways.
-
Tumor-Suppressive miRNAs: conversely, these miRNAs are downregulated in breast cancer and typically act to inhibit oncogenes or other pro-tumorigenic processes.
To maintain normal cellular function, it is essential to maintain a balance between oncogenic and tumor-suppressive miRNAs as reviewed earlier [24]. In breast cancer, dysregulation of miRNA expression contributes to tumorigenesis (Table 1). Therapeutic potential exists in manipulating miRNA expression. OncomiRs can be inhibited or replaced with miRNAs (for tumor-suppressive miRNAs).
Table 1. List of miRNAs and the function of their targets in breast cancer.
.
| MicroRNA | Target | Function | References |
|---|---|---|---|
| Oncogenic miRNAs in breast cancer | |||
| miRNA-10b | HOXD10 | Promotes cell migration, invasion, and metastasis | [25] |
| miRNA-17/92 cluster | COL4A3, LAMA3, TIMP2/3, ADORA1 |
Promotes lymph node metastasis, enhanced cell proliferation, colony formation, migration, and invasion in Triple Negative Breast Cancer (TNBC) | [26,27] |
| miRNA-21 | PDCD4, PTEN, TPM1, TIMP3 |
Promotes invasion, metastasis, and migration | [28,29,30,31] |
| miRNA-24 | Nanog, Oct-3/4, BimL, F1H1, HIF-1, Snail, VEGFA | Hypoxia-inducible miRNA | [32] |
| miRNA-122 | Pyruvate kinase and citrate synthase |
Promotes metastasis by reprogrammed glucose metabolism | [33] |
| miRNA-135b | LATS2, CDK2, p-YAP |
Promotes cell proliferation and S-G2/M cell cycle progression | [34] |
| miRNA-155 | SOCS1, TP53INP1, FOXO3 | Promotes cell growth, proliferation, and survival | [35,36,37] |
| miRNA-181a | Bim | Promotes epithelial-to-mesenchymal transition (EMT), migration, and invasion | [38] |
| miRNA-191-5p | SOX4, caspase-3, caspase-7, p53 | Promotes apoptosis resistance and doxorubicin resistance | [39] |
| miRNA-200b | Ezrin/Radixin/Moesin (ERM) | Promotes metastasis and invasion | [40] |
| miRNA-206 | NK-1 | Promotes breast cancer cell invasion, migration, proliferation, and colony formation in vitro | [41] |
| miRNA-210 | Pax-5 | Modulating EMT and hypoxia | [42] |
| miRNA-331 | HER2, HOTAIR, E2F1, DOHH |
Promotes metastasis and invasion by elevation in plasma of metastatic breast cancer patients | [43] |
| miRNA-373 | CD44 | Promotes cell migration, invasion, and metastasis | [44] |
| miRNA-455-3p | EI24 | Promotes proliferation, invasion, and migration | [45] |
| miRNA-498 | BRCA1 | Promotes TNBC cell proliferation | [46] |
| miRNA-520c | CD44 | Promotes cell migration, invasion, and metastasis | [44] |
| Tumor-suppressor miRNAs in breast cancer | |||
| miRNA-17-92 | Mekk2, cyclin D1 | Promotes NK cell antitumoral activity and reduces metastasis, regulates G1 to S phase transition | [47,48] |
| miRNA-7 | SETDB1 | Inhibits cell invasion and metastasis, decreases the BCSC population, and partially reverses EMT | [49] |
| let-7d | Cyclin D1 | Induces stem cells radiation sensitization | [50] |
| miRNA-30 | Ubc9, ITGB3 | Inhibits self-renewal of breast tumor-initiating cells (BT-ICs), trigger apoptosis | [51] |
| miRNA-33b | HMGA2, SALL4, Twist1 | Regulates cell stemness and metastasis | [52] |
| miRNA-34a | IMP3 | Regulates TNBC stem cell property | [53] |
| miRNA-125b | ERBB2, EPO, EPOR, ENPEP, CK2-α, CCNJ, MEGF9 | Inhibits cell proliferation and differentiation, migration and invasion |
[54,55] |
| miRNA-137 | FSTL1 | Suppresses TNBC stemness | [56] |
| miRNA-143 | ERK5, MAP3K7, Cyclin D1 | Anti-proliferative | [57] |
| miRNA-148a | WNT1, MMP13 | Inhibits cell proliferation, migration and invasion |
[58,59] |
| miRNA-200a | TFAM | Regulates breast cancer cell growth and mtDNA copy number | [60] |
| miRNA-203 | ΔNp63α | Forfeiture of self-renewing capacity associated with epithelial stem cells, suppresses proliferation and colony formation | [61] |
| miRNA-205 | HMGB3, Notch-2 | Suppresses proliferation and invasion and inhibits EMT and stem cell properties | [62,63] |
| miRNA-206 | Cyclin D2, Cx43 | Reduces migration, invasion, and metastasis | [64,65] |
| miRNA-223 | HAX-1 | Re-sensitizes TNBC stem cells to tumor necrosis factor-related apoptosis | [66] |
| miRNA-483-3p | Cyclin E1, p-NPAT, NPAT, CDK2 |
Anti-proliferative and G1-S cell cycle arrest | [67] |
| miRNA-497 | Cyclin E1 | Anti-proliferative and reduces migration | [68] |
| miRNA-519d-3p | LIMK1 | Suppresses growth and motility | [69] |
Studying OncomiRs and tumor-suppressive miRNAs in breast cancer is of paramount importance due to several reasons. Interactions between OncomiRs and tumor-suppressive miRNAs provide insight into the molecular mechanisms underlying breast cancer progression. The level of dysregulation of these miRNAs in tumor tissue or bodily fluids correlates with the cancer stage, aggressiveness, and clinical outcome. It is possible to tailor personalized treatment strategies for patients by identifying specific OncomiRs and tumor-suppressive miRNAs associated with breast cancer subtypes or treatment responses.
4. Significance of miRNAs in Breast Cancer Development
miRNAs function by binding to the 3′ UTR of target mRNAs, leading to translational repression or mRNA degradation. Breast cancer is characterized by dysregulation of miRNAs and their targets, which contribute to various aspects of cancer initiation, progression, and metastasis. Below is an overview of some key miRNAs and their known roles in the regulation of their targets in breast cancer.
4.1. Breast Cancer Initiation and Progression
As a multistep process, cancer initiates and progresses with the gradual transformation of human cells into highly malignant forms through progressive genetic changes [70]. It has been recognized that malignant transformation occurs through successive mutations in specific genes, leading to the activation of oncogenes and the inactivation of tumor-suppressor genes. The activities of these genes may represent the final common pathway using which many carcinogens act [71]. There are three main types of genes that play a role in tumor initiation: proto-oncogenes, tumor-suppressor genes, and genes involved in DNA repair. Changes in the genes, like mutations, amplifications, or deletions, may lead to decoupling of biological mechanism involved in the regulation of normal cell growth and differentiation [72].
Accumulating evidence suggests that cancer stem cells (CSCs) or tumor-initiating cells (TICs) with stem cell-like properties can propagate human tumors with heterogeneous tumor populations in immunodeficient mice, such as human-breast-tumor-initiating cells (BTICs) or breast cancer stem cells (BCSCs) that can regenerate breast tumors in an in vivo model. Additionally, BTICs self-renew and asymmetrically divide into differentiated cancer cells, and these are trusted to be accountable for cancer stem-like cells that drive breast tumor formation, recurrence, metastasis, and drug resistance [66,73]. Liu et al. proved that BCSCs are involved in the spontaneous metastases of human breast cancer in mouse breast cancer orthotopic models [74]. Several miRNAs have been implicated in the regulation of CSC properties. The dysregulation of miRNA might contribute to the self-renewal of BCSCs and cancer progression.
The pivotal roles of miRNA-200 are well characterized in breast cancer initiation. For example, miRNA-200 family members are significantly downregulated in CD44+, CD24−low-lineage human primary BCSCs when compared to non-tumorigenic cancer cells. Moreover, miRNA-200b regulates BCSC growth by directly targeting Suz12, a subunit of a polycomb repressor complex (PRC2) and regulates EMT by repressing the E-cadherin gene. Higher expression of the polycomb protein EZH2, which is important in the stem cell self-renewal capability of embryonic and adult stem cells, has been associated with breast cancer progression [75]. Shimono et al. reported that the expression of the tumor-suppressor miRNA-200c decreased the self-renewal ability of BCSCs in vitro and tumor formation ability in vivo [76].
Yu et al. showed that let-7 and miRNA-30e were downregulated in BCSCs and SK-3rd mammospheres compared to non-tumorigenic cells or more differentiated cells, respectively. Enforced expression of miRNA-30e inhibited mammosphere formation and tumorigenesis of SK-3rd cells in vitro and in vivo, respectively, by targeting ITGB3 and UBC9 [51]. The miRNA-128 was found to be downregulated in BCSCs (in CD44+, CD24−/low, and mammospheres) compared to non-cancerous cells [77]. Furthermore, miRNA-128 is significantly low in chemoresistant BTICs enriched from breast cancer cells (SK-3rd and MCF-7) and primary breast tumors, and that its target Bmi-1 and ABCC5 are overexpressed in these cancers. The ectopic expression of miRNA-128 sensitizes BTICs to the proapoptotic and DNA-damaging effects of doxorubicin, showing its therapeutic potential [77]. In another finding, miRNA-30c regulates invasion of breast cancer by targeting the cytoskeleton network genes encoding Twinfilin 1 (TWF1) and Vimentin (VIM), both of which regulate EMT [78].
The miRNA-34c expression level and function were reduced in the BTICs of MCF-7 and SK-3rd cells, and these cell lines were enriched for BTICs [79]. When ectopic expression of miRNA-34c decreased the self-renewal of BTICs, inhibited EMT, and suppressed migration of the tumor cells through inhibition of the target gene Notch4, miRNA-495 was significantly upregulated in CD44+/CD24−/low BCSCs, reflecting its potential importance in maintaining common BCSC properties, and its ectopic expression promoted colony formation in vitro and tumorigenesis in mice. In addition, the overexpression of miRNA-495 is associated with the lowered expression of E-cadherin, which enhances the stem-like phenotype in BCSCs [80]. miRNA-181 family members and miRNA-155 oncogenic miRNAs promote self-renewal, sphere formation, colony formation, or tumor development in breast cancer cells [81].
4.2. miRNA in Metastatic Breast Cancer
Metastatic breast cancer is a complex, multistep malignant process through which tumor cells migrate from their primary tumor (site of origin) to colonize distant tissues (e.g., liver, brain, bones, or lungs) and is often responsible for 90% of cancer-related mortality [82,83]. EMT is one of the important processes by which cancer metastasis starts, and EMT induces morphological changes in epithelial cells by which epithelial cells transform into mesenchymal cells. During this process, cancer cells lose their cell–cell communication and become mobile and invasive, spreading into distant organs and tissues [84]. Suppression of E-cadherin expression in epithelial cancer cells is a hallmark of EMT. Various miRNAs have been linked to the control of EMT in cancer, and several genes, including ZEB [85], Twist [86], Snail [87], and Slug [88] are known to restrict the expression of E-cadherin.
One of the most important miRNAs that control metastasis is the miRNA-200 family (miRNA-200a/200b/200c/141/429), and this prevents cell migration and invasion by targeting ZEB in several cancer types, including breast cancer [89], and miRNA-200/ZEB plays a central role in the EMT/MET processes. In the meantime, inhibition of miRNA-200 reduces the E-cadherin level while supporting VIM expression, thereby increasing cell motility [89]. Many studies also demonstrated that TGF-β-induced EMT might be inhibited only with the ectopic expression of the miRNA-200 family [90]. Xie et al. reported that miRNA-193a-WT1 interaction plays an important role in breast cancer metastasis and indicates that restoring miRNA-193a expression is a therapeutic strategy in breast cancer [91].
Ma and co-workers discovered that upregulation of miRNA-10b promotes invasion and metastasis by indirectly activating the pro-metastatic gene RHOC through repressing HOXD10 [25]. In addition, miRNA-373 regulates the CD44 gene, which promotes tumor invasion and metastasis [44]. miRNA-9, a MYC/MYCN-induced miRNA, has been demonstrated to directly target E-cadherin to promote breast cancer metastasis via activation of β-catenin signaling, through increasing tumor invasiveness and angiogenesis [92].
Recently, several studies have reported that Dicer, an endonuclease that processes miRNAs, is also associated with EMT and metastatic progression. Dicer inhibition promotes metastasis, and restoring its expression suppresses metastasis. Certain miRNAs also target Dicer for controlling metastasis. For example, miRNA-103/107 induced EMT by targeting Dicer expression [93]. In addition, the miRNA-221/222 cluster has been shown to induce EMT in breast cancer cells by targeting Dicer, and estrogen receptor 1 (ESR1) [94]. In breast cancer, miRNA-107 acts as an endogenous suppressor of let-7, and it interacts with mature let-7 to inhibit its function, which plays a major role in determining metastatic progression [95]. According to Huang et al., both in vitro and in vivo cancer cell migration and invasion were promoted by miRNA-373 and miRNA-520c [44]. Wu et al. reported that in breast cancer cells, miRNA-29a regulates the critical roles of EMT and metastasis by targeting SUV420H2 [96].
The overexpression of miRNA-17/92 is also involved in metastatic breast cancer [27]. miRNA-454-3p plays an important role in breast cancer’s early metastatic events, promotes the stemness of breast cancer cells, and promotes early distant relapse in both in vitro and in vivo conditions. The higher expression of miRNA-454-3p was found to be significantly associated with both a poor prognosis and early recurrence in breast cancer through Wnt/β-catenin signaling activation [97]. Higher expression of miRNA-373 was found in breast cancer samples from tumors exhibiting lymph node metastasis [44]. Dobson et al. identified a novel target of miRNA-30c, the nephroblastoma overexpressed gene (NOV), which is an inhibitor of the invasiveness of metastatic TNBC (MDA-MB-231) cells [98]. The miRNA-34a plays a key role in the proliferation, invasion, and metastasis of breast cancer cells [53]. Moreover, miRNA-373, miRNA-520c, miRNA-210, and miRNA-29b were also shown to influence the invasion and migration of breast cancer [44]. In addition, oncogenic miRNA-224 expression is significantly upregulated in highly invasive MDA-MB-231 cells and correlated with increased metastasis [99]. Another important oncogenic miRNA, miRNA-155, is frequently overexpressed in invasive breast cancer tissues and is directly targeted to RhoA and contributes to breast cancer metastasis. Inhibition of miRNA-155 suppressed TGF-β-induced EMT and tight junction dissolution, along with cell migration and invasion. Further, the ectopic expression of miRNA-155 reduced RhoA protein and disrupted tight junction formation [100]. miRNA-21 is an important oncogene and has a role in breast cancer tumorigenesis. It regulates invasion and tumor metastasis by targeting multiple tumor and metastasis suppressor genes. Also, inhibiting miRNA-21 reduced the invasion and lung metastasis of MDA-MB-231 cells [30]. All these studies demonstrated that some of the miRNAs have a metastatic role in breast cancer cells.
This entry is adapted from the peer-reviewed paper 10.3390/biomedicines12030691
This entry is offline, you can click here to edit this entry!
