The ORF3a (open reading frame 3a) protein found in SARS-CoV-2, represents a promising target for antiviral treatment due to its multifaceted role in viral pathogenesis, cytokine storms, disease severity, and mortality. ORF3a contributes significantly to viral pathogenesis by facilitating viral assembly and release, essential processes in the viral life cycle, while also suppressing the body’s antiviral responses, thus aiding viral replication. ORF3a also has been implicated in triggering excessive inflammation, characterized by NF-κB-mediated cytokine production, ultimately leading to apoptotic cell death and tissue damage in the lungs, kidneys, and the central nervous system.
- SARS-CoV-2
- COVID-19
- ORF3a
1. Introduction
2. SARS-CoV-2 ORF3a Protein as a Promising Antiviral Target
3. Structure and Unique Features of ORF3a Protein
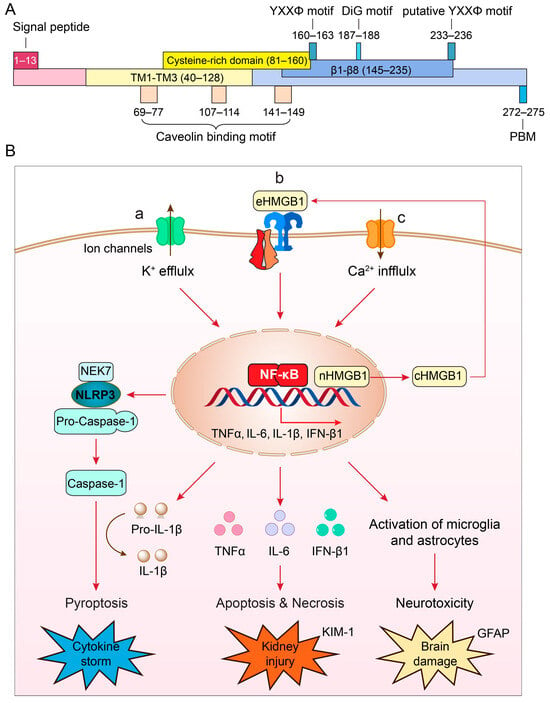
4. Role of ORF3a in Viral Production
5. Role of ORF3a in Viral Pathogenesis: Induction of Cytokine Storm and COVID-19 Severity
6. Roles of ORF3a in Kidney Injury, Neuroinflammation, and Long-COVID Complications
7. Discovery and Development of Inhibitor Drugs Targeting ORF3a Protein
7.1. Testing Well-Established Druggable Cellular Targets of Key ORF3a Regulators with FDA-Approved Drugs and Small Molecule Inhibitors (SMIs)
7.2. Structure-Based Design of ORF3a Inhibitors
7.3. Cell-Based High-Throughput Drug Screening
This entry is adapted from the peer-reviewed paper 10.3390/pathogens13010075
References
- Alcendor, D.J.; Matthews-Juarez, P.; Smoot, D.; Hildreth, J.E.K.; Lamar, K.; Tabatabai, M.; Wilus, D.; Juarez, P.D. Breakthrough COVID-19 Infections in the US: Implications for Prolonging the Pandemic. Vaccines 2022, 10, 755.
- Rubin, R. From Positive to Negative to Positive Again-The Mystery of Why COVID-19 Rebounds in Some Patients Who Take Paxlovid. JAMA 2022, 327, 2380–2382.
- Service, R.F. Bad news for Paxlovid? Resistance may be coming. Science 2022, 377, 138–139.
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of COVID-19—Final Report. N. Engl. J. Med. 2020, 383, 1813–1826.
- Zhao, R.Y. Yeast for virus research. Microb. Cell 2017, 4, 311–330.
- Li, G.; Zhao, R.Y. Molecular Cloning and Characterization of Small Viral Genome in Fission Yeast. Methods Mol. Biol. 2018, 1721, 47–61.
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468.
- Harcourt, J.; Tamin, A.; Lu, X.; Kamili, S.; Sakthivel, S.K.; Murray, J.; Queen, K.; Tao, Y.; Paden, C.R.; Zhang, J.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with Coronavirus Disease, United States. Emerg. Infect. Dis. 2020, 26, 1266–1273.
- Zhao, Y.; Elder, R.T.; Chen, M.; Cao, J. Fission yeast expression vectors adapted for positive identification of gene insertion and green fluorescent protein fusion. Biotechniques 1998, 25, 438–440, 442, 444.
- Nkeze, J.; Li, L.; Benko, Z.; Li, G.; Zhao, R.Y. Molecular characterization of HIV-1 genome in fission yeast Schizosaccharomyces pombe. Cell Biosci. 2015, 5, 47.
- Li, G.; Poulsen, M.; Fenyvuesvolgyi, C.; Yashiroda, Y.; Yoshida, M.; Simard, J.M.; Gallo, R.C.; Zhao, R.Y. Characterization of cytopathic factors through genome-wide analysis of the Zika viral proteins in fission yeast. Proc. Natl. Acad. Sci. USA 2017, 114, E376–E385.
- Zhang, J.; Li, Q.; Cruz Cosme, R.S.; Gerzanich, V.; Tang, Q.; Simard, J.M.; Zhao, R.Y. Genome-Wide Characterization of SARS-CoV-2 Cytopathogenic Proteins in the Search of Antiviral Targets. mBio 2021, 13, e0016922.
- Zhang, J.; Ejikemeuwa, A.; Gerzanich, V.; Nasr, M.; Tang, Q.; Simard, J.M.; Zhao, R.Y. Understanding the Role of SARS-CoV-2 ORF3a in Viral Pathogenesis and COVID-19. Front. Microbiol. 2022, 13, 854567.
- Hachim, A.; Kavian, N.; Cohen, C.A.; Chin, A.W.H.; Chu, D.K.W.; Mok, C.K.P.; Tsang, O.T.Y.; Yeung, Y.C.; Perera, R.; Poon, L.L.M.; et al. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020, 21, 1293–1301.
- Camerini, D.; Randall, A.Z.; Trappl-Kimmons, K.; Oberai, A.; Hung, C.; Edgar, J.; Shandling, A.; Huynh, V.; Teng, A.A.; Hermanson, G.; et al. Mapping SARS-CoV-2 Antibody Epitopes in COVID-19 Patients with a Multi-Coronavirus Protein Microarray. Microbiol. Spectr. 2021, 9, e0141621.
- Liu, I.J.; Hsueh, P.R.; Lin, C.T.; Chiu, C.Y.; Kao, C.L.; Liao, M.Y.; Wu, H.C. Disease-specific B Cell epitopes for serum antibodies from patients with severe acute respiratory syndrome (SARS) and serologic detection of SARS antibodies by epitope-based peptide antigens. J. Infect. Dis. 2004, 190, 797–809.
- Zhong, X.; Guo, Z.; Yang, H.; Peng, L.; Xie, Y.; Wong, T.Y.; Lai, S.T.; Guo, Z. Amino terminus of the SARS coronavirus protein 3a elicits strong, potentially protective humoral responses in infected patients. J. Gen. Virol. 2006, 87, 369–373.
- Zhu, A.; Liu, M.; Li, Y.; Lei, Q.; Wu, Q.; Lin, M.; Lai, D.; Lu, L.; Yu, S.; Guo, S.; et al. Age- and Severity-Associated Humoral Immunity Response in COVID-19 Patients: A Cohort Study from Wuhan, China. J. Clin. Med. 2022, 11, 5974.
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e1415.
- Tan, A.T.; Linster, M.; Tan, C.W.; Le Bert, N.; Chia, W.N.; Kunasegaran, K.; Zhuang, Y.; Tham, C.Y.L.; Chia, A.; Smith, G.J.D.; et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021, 34, 108728.
- Samandari, T.; Ongalo, J.B.; McCarthy, K.D.; Biegon, R.K.; Madiega, P.A.; Mithika, A.; Orinda, J.; Mboya, G.M.; Mwaura, P.; Anzala, O.; et al. Prevalence and functional profile of SARS-CoV-2 T cells in asymptomatic Kenyan adults. J. Clin. Investig. 2023, 133, 13.
- Kern, D.M.; Sorum, B.; Mali, S.S.; Hoel, C.M.; Sridharan, S.; Remis, J.P.; Toso, D.B.; Kotecha, A.; Bautista, D.M.; Brohawn, S.G. Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. Nat. Struct. Mol. Biol. 2021, 28, 573–582.
- McClenaghan, C.; Hanson, A.; Lee, S.J.; Nichols, C.G. Coronavirus Proteins as Ion Channels: Current and Potential Research. Front. Immunol. 2020, 11, 573339.
- Issa, E.; Merhi, G.; Panossian, B.; Salloum, T.; Tokajian, S. SARS-CoV-2 and ORF3a: Nonsynonymous Mutations, Functional Domains, and Viral Pathogenesis. mSystems 2020, 5, e00266-20.
- Cruz-Cosme, R.; Zhang, J.; Liu, D.; Mahase, V.; Sallapalli, B.T.; Chang, P.; Zhang, Y.; Teng, S.; Zhao, R.Y.; Tang, Q. A novel diG motif in ORF3a protein of SARS-CoV-2 for intracellular transport. Front. Cell Dev. Biol. 2022, 10, 1011221.
- Padhan, K.; Tanwar, C.; Hussain, A.; Hui, P.Y.; Lee, M.Y.; Cheung, C.Y.; Peiris, J.S.M.; Jameel, S. Severe acute respiratory syndrome coronavirus Orf3a protein interacts with caveolin. J. Gen. Virol. 2007, 88, 3067–3077.
- Tan, Y.J.; Teng, E.; Shen, S.; Tan, T.H.; Goh, P.Y.; Fielding, B.C.; Ooi, E.E.; Tan, H.C.; Lim, S.G.; Hong, W. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004, 78, 6723–6734.
- Minakshi, R.; Padhan, K. The YXXPhi motif within the severe acute respiratory syndrome coronavirus (SARS-CoV) 3a protein is crucial for its intracellular transport. Virol. J. 2014, 11, 75.
- Bianchi, M.; Borsetti, A.; Ciccozzi, M.; Pascarella, S. SARS-CoV-2 ORF3a: Mutability and function. Int. J. Biol. Macromol. 2021, 170, 820–826.
- Kakkanas, A.; Karamichali, E.; Koufogeorgou, E.I.; Kotsakis, S.D.; Georgopoulou, U.; Foka, P. Targeting the YXXPhi Motifs of the SARS Coronaviruses 1 and 2 ORF3a Peptides by In Silico Analysis to Predict Novel Virus-Host Interactions. Biomolecules 2022, 12, 1052.
- Siu, K.L.; Yuen, K.S.; Castano-Rodriguez, C.; Ye, Z.W.; Yeung, M.L.; Fung, S.Y.; Yuan, S.; Chan, C.P.; Yuen, K.Y.; Enjuanes, L.; et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019, 33, 8865–8877.
- Jin, D.Y.; Zheng, B.J.; Tang, H.M.V. Mechanism of inflammasome activation by SARS coronavirus 3a protein: Abridged secondary publication. Hong Kong Med. J. 2021, 27 (Suppl. S2), 33–35.
- Guarnieri, J.W.; Angelin, A.; Murdock, D.G.; Schaefer, P.; Portluri, P.; Lie, T.; Huang, J.; Wallace, D.C. SARS-CoV-2 viroporins activate the NLRP3-inflammasome by the mitochondrial permeability transition pore. Front. Immunol. 2023, 14, 1064293.
- Cai, H.; Chen, Y.; Feng, Y.; Asadi, M.; Kaufman, L.; Lee, K.; Kehrer, T.; Miorin, L.; Garcia-Sastre, A.; Gusella, G.L.; et al. SARS-CoV-2 viral protein ORF3A injures renal tubules by interacting with TRIM59 to induce STAT3 activation. Mol. Ther. 2023, 31, 774–787.
- Zhu, H.; Byrnes, C.; Lee, Y.T.; Tuymetova, G.; Duffy, H.B.D.; Bakir, J.Y.; Pettit, S.N.; Angina, J.; Springer, D.A.; Allende, M.L.; et al. SARS-CoV-2 ORF3a expression in brain disrupts the autophagy-lysosomal pathway, impairs sphingolipid homeostasis, and drives neuropathogenesis. FASEB J. 2023, 37, e22919.
- Han, W.K.; Bailly, V.; Abichandani, R.; Thadhani, R.; Bonventre, J.V. Kidney Injury Molecule-1 (KIM-1): A novel biomarker for human renal proximal tubule injury. Kidney Int. 2002, 62, 237–244.
- Liddelow, S.A.; Guttenplan, K.A.; Clarke, L.E.; Bennett, F.C.; Bohlen, C.J.; Schirmer, L.; Bennett, M.L.; Munch, A.E.; Chung, W.S.; Peterson, T.C.; et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017, 541, 481–487.
- Schiff, L.; Hadker, N.; Weiser, S.; Rausch, C. A literature review of the feasibility of glial fibrillary acidic protein as a biomarker for stroke and traumatic brain injury. Mol. Diagn. Ther. 2012, 16, 79–92.
- Brant, A.C.; Tian, W.; Majerciak, V.; Yang, W.; Zheng, Z.M. SARS-CoV-2: From its discovery to genome structure, transcription, and replication. Cell Biosci. 2021, 11, 136.
- Lee, Y.B.; Jung, M.; Kim, J.; Charles, A.; Christ, W.; Kang, J.; Kang, M.G.; Kwak, C.; Klingstrom, J.; Smed-Sorensen, A.; et al. Super-resolution proximity labeling reveals anti-viral protein network and its structural changes against SARS-CoV-2 viral proteins. Cell Rep. 2023, 42, 112835.
- Breitinger, U.; Farag, N.S.; Sticht, H.; Breitinger, H.G. Viroporins: Structure, function, and their role in the life cycle of SARS-CoV-2. Int. J. Biochem. Cell Biol. 2022, 145, 106185.
- Xu, H.; Akinyemi, I.A.; Chitre, S.A.; Loeb, J.C.; Lednicky, J.A.; McIntosh, M.T.; Bhaduri-McIntosh, S. SARS-CoV-2 viroporin encoded by ORF3a triggers the NLRP3 inflammatory pathway. Virology 2022, 568, 13–22.
- Fam, M.S.; Sedky, C.A.; Turky, N.O.; Breitinger, H.G.; Breitinger, U. Channel activity of SARS-CoV-2 viroporin ORF3a inhibited by adamantanes and phenolic plant metabolites. Sci. Rep. 2023, 13, 5328.
- Nieva, J.L.; Madan, V.; Carrasco, L. Viroporins: Structure and biological functions. Nat. Rev. Microbiol. 2012, 10, 563–574.
- Zhang, J.; Cruz-Cosme, R.; Zhang, C.; Liu, D.; Tang, Q.; Zhao, R.Y. Endoplasmic reticulum-associated SARS-CoV-2 ORF3a elicits heightened cytopathic effects despite robust ER-associated degradation. mBio 2023, e0303023.
- Busscher, B.M.; Befekadu, H.B.; Liu, Z.; Xiao, T.S. SARS-CoV-2 ORF3a-Mediated NF-kappaB Activation Is Not Dependent on TRAF-Binding Sequence. Viruses 2023, 15, 2229.
- Caillet-Saguy, C.; Durbesson, F.; Rezelj, V.V.; Gogl, G.; Tran, Q.D.; Twizere, J.C.; Vignuzzi, M.; Vincentelli, R.; Wolff, N. Host PDZ-containing proteins targeted by SARS-CoV-2. FEBS J. 2021, 288, 5148–5162.
- Castano-Rodriguez, C.; Honrubia, J.M.; Gutierrez-Alvarez, J.; DeDiego, M.L.; Nieto-Torres, J.L.; Jimenez-Guardeno, J.M.; Regla-Nava, J.A.; Fernandez-Delgado, R.; Verdia-Baguena, C.; Queralt-Martin, M.; et al. Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis. mBio 2018, 9, 10–1128.
- Rice, A.P.; Kimata, J.T. SARS-CoV-2 likely targets cellular PDZ proteins: A common tactic of pathogenic viruses. Future Virol. 2021, 16, 375–377.
- Chen, D.; Zheng, Q.; Sun, L.; Ji, M.; Li, Y.; Deng, H.; Zhang, H. ORF3a of SARS-CoV-2 promotes lysosomal exocytosis-mediated viral egress. Dev. Cell 2021, 56, 3250–3263.e5.
- Lu, W.; Zheng, B.J.; Xu, K.; Schwarz, W.; Du, L.; Wong, C.K.; Chen, J.; Duan, S.; Deubel, V.; Sun, B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. USA 2006, 103, 12540–12545.
- Ghosh, S.; Dellibovi-Ragheb, T.A.; Kerviel, A.; Pak, E.; Qiu, Q.; Fisher, M.; Takvorian, P.M.; Bleck, C.; Hsu, V.W.; Fehr, A.R.; et al. beta-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell 2020, 183, 1520–1535.e1514.
- Yount, B.; Roberts, R.S.; Sims, A.C.; Deming, D.; Frieman, M.B.; Sparks, J.; Denison, M.R.; Davis, N.; Baric, R.S. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005, 79, 14909–14922.
- Akerstrom, S.; Mirazimi, A.; Tan, Y.J. Inhibition of SARS-CoV replication cycle by small interference RNAs silencing specific SARS proteins, 7a/7b, 3a/3b and S. Antivir. Res. 2007, 73, 219–227.
- Silvas, J.A.; Vasquez, D.M.; Park, J.G.; Chiem, K.; Allue-Guardia, A.; Garcia-Vilanova, A.; Platt, R.N.; Miorin, L.; Kehrer, T.; Cupic, A.; et al. Contribution of SARS-CoV-2 Accessory Proteins to Viral Pathogenicity in K18 Human ACE2 Transgenic Mice. J. Virol. 2021, 95, e0040221.
- Liu, Y.; Zhang, X.; Liu, J.; Xia, H.; Zou, J.; Muruato, A.E.; Periasamy, S.; Kurhade, C.; Plante, J.A.; Bopp, N.E.; et al. A live-attenuated SARS-CoV-2 vaccine candidate with accessory protein deletions. Nat. Commun. 2022, 13, 4337.
- Duarte, L.F.; Vazquez, Y.; Diethelm-Varela, B.; Pavez, V.; Berrios-Rojas, R.; Mendez, C.; Riedel, C.A.; White, J.A.; Kalergis, A.M.; Bueno, S.M.; et al. Differential Severe Acute Respiratory Syndrome Coronavirus 2-Specific Humoral Response in Inactivated Virus-Vaccinated, Convalescent, and Breakthrough-Infected Subjects. J. Infect. Dis. 2023, 228, 857–867.
- Zhang, J.; Wu, H.; Yao, X.; Zhang, D.; Zhou, Y.; Fu, B.; Wang, W.; Li, H.; Wang, Z.; Hu, Z.; et al. Pyroptotic macrophages stimulate the SARS-CoV-2-associated cytokine storm. Cell Mol. Immunol. 2021, 18, 1305–1307.
- Ratajczak, M.Z.; Kucia, M. SARS-CoV-2 infection and overactivation of Nlrp3 inflammasome as a trigger of cytokine “storm” and risk factor for damage of hematopoietic stem cells. Leukemia 2020, 34, 1726–1729.
- Hu, B.; Huang, S.; Yin, L. The cytokine storm and COVID-19. J. Med. Virol. 2021, 93, 250–256.
- Freeman, T.L.; Swartz, T.H. Targeting the NLRP3 Inflammasome in Severe COVID-19. Front. Immunol. 2020, 11, 1518.
- Van den Berg, D.F.; Te Velde, A.A. Severe COVID-19: NLRP3 Inflammasome Dysregulated. Front. Immunol. 2020, 11, 1580.
- Bertoni, A.; Penco, F.; Mollica, H.; Bocca, P.; Prigione, I.; Corcione, A.; Cangelosi, D.; Schena, F.; Del Zotto, G.; Amaro, A.; et al. Spontaneous NLRP3 inflammasome-driven IL1-beta secretion is induced in severe COVID-19 patients and responds to anakinra treatment. J. Allergy Clin. Immunol. 2022, 150, 796–805.
- Jorgensen, I.; Miao, E.A. Pyroptotic cell death defends against intracellular pathogens. Immunol. Rev. 2015, 265, 130–142.
- Yang, Y.; Wang, H.; Kouadir, M.; Song, H.; Shi, F. Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death Dis. 2019, 10, 128.
- Yang, H.; Wang, H.; Andersson, U. Targeting Inflammation Driven by HMGB1. Front. Immunol. 2020, 11, 484.
- Gowda, P.; Patrick, S.; Joshi, S.D.; Kumawat, R.K.; Sen, E. Glycyrrhizin prevents SARS-CoV-2 S1 and Orf3a induced high mobility group box 1 (HMGB1) release and inhibits viral replication. Cytokine 2021, 142, 155496.
- Paudel, Y.N.; Shaikh, M.F.; Chakraborti, A.; Kumari, Y.; Aledo-Serrano, A.; Aleksovska, K.; Alvim, M.K.M.; Othman, I. HMGB1: A Common Biomarker and Potential Target for TBI, Neuroinflammation, Epilepsy, and Cognitive Dysfunction. Front. Neurosci. 2018, 12, 628.
- Peng, T.; Du, S.Y.; Son, M.; Diamond, B. HIF-1alpha is a negative regulator of interferon regulatory factors: Implications for interferon production by hypoxic monocytes. Proc. Natl. Acad. Sci. USA 2021, 118, e2106017118.
- Bolay, H.; Karadas, O.; Ozturk, B.; Sonkaya, R.; Tasdelen, B.; Bulut, T.D.S.; Gulbahar, O.; Ozge, A.; Baykan, B. HMGB1, NLRP3, IL-6 and ACE2 levels are elevated in COVID-19 with headache: A window to the infection-related headache mechanism. J. Headache Pain 2021, 22, 94.
- Chen, L.; Long, X.; Xu, Q.; Tan, J.; Wang, G.; Cao, Y.; Wei, J.; Luo, H.; Zhu, H.; Huang, L.; et al. Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients. Cell Mol. Immunol. 2020, 17, 992–994.
- Legrand, M.; Bell, S.; Forni, L.; Joannidis, M.; Koyner, J.L.; Liu, K.; Cantaluppi, V. Pathophysiology of COVID-19-associated acute kidney injury. Nat. Rev. Nephrol. 2021, 17, 751–764.
- May, R.M.; Cassol, C.; Hannoudi, A.; Larsen, C.P.; Lerma, E.V.; Haun, R.S.; Braga, J.R.; Hassen, S.I.; Wilson, J.; VanBeek, C.; et al. A multi-center retrospective cohort study defines the spectrum of kidney pathology in Coronavirus 2019 Disease (COVID-19). Kidney Int. 2021, 100, 1303–1315.
- Bowe, B.; Cai, M.; Xie, Y.; Gibson, A.K.; Maddukuri, G.; Al-Aly, Z. Acute Kidney Injury in a National Cohort of Hospitalized US Veterans with COVID-19. Clin. J. Am. Soc. Nephrol. 2020, 16, 14–25.
- Chan, L.; Chaudhary, K.; Saha, A.; Chauhan, K.; Vaid, A.; Zhao, S.; Paranjpe, I.; Somani, S.; Richter, F.; Miotto, R.; et al. AKI in Hospitalized Patients with COVID-19. J. Am. Soc. Nephrol. 2021, 32, 151–160.
- Geri, G.; Darmon, M.; Zafrani, L.; Fartoukh, M.; Voiriot, G.; Le Marec, J.; Nemlaghi, S.; Vieillard-Baron, A.; Azoulay, E. Acute kidney injury in SARS-CoV2-related pneumonia ICU patients: A retrospective multicenter study. Ann. Intensive Care 2021, 11, 86.
- Amraei, R.; Yin, W.; Napoleon, M.A.; Suder, E.L.; Berrigan, J.; Zhao, Q.; Olejnik, J.; Chandler, K.B.; Xia, C.; Feldman, J.; et al. CD209L/L-SIGN and CD209/DC-SIGN Act as Receptors for SARS-CoV-2. ACS Cent. Sci. 2021, 7, 1156–1165.
- Kalejaiye, T.D.; Bhattacharya, R.; Burt, M.A.; Travieso, T.; Okafor, A.E.; Mou, X.; Blasi, M.; Musah, S. SARS-CoV-2 Employ BSG/CD147 and ACE2 Receptors to Directly Infect Human Induced Pluripotent Stem Cell-Derived Kidney Podocytes. Front. Cell Dev. Biol. 2022, 10, 855340.
- Khan, S.; Chen, L.; Yang, C.R.; Raghuram, V.; Khundmiri, S.J.; Knepper, M.A. Does SARS-CoV-2 Infect the Kidney? J. Am. Soc. Nephrol. 2020, 31, 2746–2748.
- Armaly, Z.; Kinaneh, S.; Skorecki, K. Renal Manifestations of COVID-19: Physiology and Pathophysiology. J. Clin. Med. 2021, 10, 1216.
- Puelles, V.G.; Lutgehetmann, M.; Lindenmeyer, M.T.; Sperhake, J.P.; Wong, M.N.; Allweiss, L.; Chilla, S.; Heinemann, A.; Wanner, N.; Liu, S.; et al. Multiorgan and Renal Tropism of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 590–592.
- Su, H.; Yang, M.; Wan, C.; Yi, L.X.; Tang, F.; Zhu, H.Y.; Yi, F.; Yang, H.C.; Fogo, A.B.; Nie, X.; et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020, 98, 219–227.
- Braun, F.; Lutgehetmann, M.; Pfefferle, S.; Wong, M.N.; Carsten, A.; Lindenmeyer, M.T.; Norz, D.; Heinrich, F.; Meissner, K.; Wichmann, D.; et al. SARS-CoV-2 renal tropism associates with acute kidney injury. Lancet 2020, 396, 597–598.
- Papadimitriou, J.C.; Drachenberg, C.B.; Kleiner, D.; Choudhri, N.; Haririan, A.; Cebotaru, V. Tubular Epithelial and Peritubular Capillary Endothelial Injury in COVID-19 AKI. Kidney Int. Rep. 2021, 6, 518–525.
- Alexander, M.P.; Mangalaparthi, K.K.; Madugundu, A.K.; Moyer, A.M.; Adam, B.A.; Mengel, M.; Singh, S.; Herrmann, S.M.; Rule, A.D.; Cheek, E.H.; et al. Acute Kidney Injury in Severe COVID-19 Has Similarities to Sepsis-Associated Kidney Injury: A Multi-Omics Study. Mayo Clin. Proc. 2021, 96, 2561–2575.
- Diao, B.; Wang, C.; Wang, R.; Feng, Z.; Zhang, J.; Yang, H.; Tan, Y.; Wang, H.; Wang, C.; Liu, L.; et al. Human kidney is a target for novel severe acute respiratory syndrome coronavirus 2 infection. Nat. Commun. 2021, 12, 2506.
- Sharfuddin, A.A.; Molitoris, B.A. Pathophysiology of ischemic acute kidney injury. Nat. Rev. Nephrol. 2011, 7, 189–200.
- Aranda-Rivera, A.K.; Cruz-Gregorio, A.; Aparicio-Trejo, O.E.; Pedraza-Chaverri, J. Mitochondrial Redox Signaling and Oxidative Stress in Kidney Diseases. Biomolecules 2021, 11, 1144.
- Guo, L.P.; Liu, S.X.; Yang, Q.; Liu, H.Y.; Xu, L.L.; Hao, Y.H.; Zhang, X.Q. Effect of Thymoquinone on Acute Kidney Injury Induced by Sepsis in BALB/c Mice. Biomed. Res. Int. 2020, 2020, 1594726.
- Meinhardt, J.; Radke, J.; Dittmayer, C.; Franz, J.; Thomas, C.; Mothes, R.; Laue, M.; Schneider, J.; Brunink, S.; Greuel, S.; et al. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 2021, 24, 168–175.
- Spudich, S.; Nath, A. Nervous system consequences of COVID-19. Science 2022, 375, 267–269.
- Crunfli, F.; Carregari, V.C.; Veras, F.P.; Silva, L.S.; Nogueira, M.H.; Antunes, A.; Vendramini, P.H.; Valenca, A.G.F.; Brandao-Teles, C.; Zuccoli, G.D.S.; et al. Morphological, cellular, and molecular basis of brain infection in COVID-19 patients. Proc. Natl. Acad. Sci. USA 2022, 119, e2200960119.
- Dedoni, S.; Avdoshina, V.; Camoglio, C.; Siddi, C.; Fratta, W.; Scherma, M.; Fadda, P. K18- and CAG-hACE2 Transgenic Mouse Models and SARS-CoV-2: Implications for Neurodegeneration Research. Molecules 2022, 27, 4142.
- Xu, E.; Xie, Y.; Al-Aly, Z. Long-term neurologic outcomes of COVID-19. Nat. Med. 2022, 28, 2406–2415.
- Marshall, M. How COVID-19 can damage the brain. Nature 2020, 585, 342–343.
- Kayagaki, N.; Stowe, I.B.; Lee, B.L.; O’Rourke, K.; Anderson, K.; Warming, S.; Cuellar, T.; Haley, B.; Roose-Girma, M.; Phung, Q.T.; et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015, 526, 666–671.
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665.
- Chen, I.Y.; Moriyama, M.; Chang, M.F.; Ichinohe, T. Severe Acute Respiratory Syndrome Coronavirus Viroporin 3a Activates the NLRP3 Inflammasome. Front. Microbiol. 2019, 10, 50.
- Bakhshi, S.; Shamsi, S. MCC950 in the treatment of NLRP3-mediated inflammatory diseases: Latest evidence and therapeutic outcomes. Int. Immunopharmacol. 2022, 106, 108595.
- He, Y.; Chang, Y.; Peng, Y.; Zhu, J.; Liu, K.; Chen, J.; Wu, Y.; Ji, Z.; Lin, Z.; Wang, S.; et al. Glibenclamide Directly Prevents Neuroinflammation by Targeting SUR1-TRPM4-Mediated NLRP3 Inflammasome Activation in Microglia. Mol. Neurobiol. 2022, 59, 6590–6607.
- Laplantine, E.; Chable-Bessia, C.; Oudin, A.; Swain, J.; Soria, A.; Merida, P.; Gourdelier, M.; Mestiri, S.; Besseghe, I.; Bremaud, E.; et al. The FDA-approved drug Auranofin has a dual inhibitory effect on SARS-CoV-2 entry and NF-kappaB signaling. iScience 2022, 25, 105066.
- Su, C.M.; Wang, L.; Yoo, D. Activation of NF-kappaB and induction of proinflammatory cytokine expressions mediated by ORF7a protein of SARS-CoV-2. Sci. Rep. 2021, 11, 13464.
- Sayah, W.; Berkane, I.; Guermache, I.; Sabri, M.; Lakhal, F.Z.; Yasmine Rahali, S.; Djidjeli, A.; Lamara Mahammed, L.; Merah, F.; Belaid, B.; et al. Interleukin-6, procalcitonin and neutrophil-to-lymphocyte ratio: Potential immune-inflammatory parameters to identify severe and fatal forms of COVID-19. Cytokine 2021, 141, 155428.
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643.
- Izadi, Z.; Brenner, E.J.; Mahil, S.K.; Dand, N.; Yiu, Z.Z.N.; Yates, M.; Ungaro, R.C.; Zhang, X.; Agrawal, M.; Colombel, J.F.; et al. Association Between Tumor Necrosis Factor Inhibitors and the Risk of Hospitalization or Death Among Patients with Immune-Mediated Inflammatory Disease and COVID-19. JAMA Netw. Open 2021, 4, e2129639.
- Lonze, B.E.; Spiegler, P.; Wesson, R.N.; Alachkar, N.; Petkova, E.; Weldon, E.P.; Dieter, R.A.; Li, Y.; Quinn, M.; Mattoo, A.; et al. A Randomized Double-Blinded Placebo Controlled Trial of Clazakizumab for the Treatment of COVID-19 Pneumonia with Hyperinflammation. Crit. Care Med. 2022, 50, 1348–1359.
- Lebedeva, N.S.; Gubarev, Y.A.; Mamardashvili, G.M.; Zaitceva, S.V.; Zdanovich, S.A.; Malyasova, A.S.; Romanenko, J.V.; Koifman, M.O.; Koifman, O.I. Theoretical and experimental study of interaction of macroheterocyclic compounds with ORF3a of SARS-CoV-2. Sci. Rep. 2021, 11, 19481.
- Arora, A.; Maiti, S. Differential biophysical behavior of human telomeric RNA and DNA quadruplex. J. Phys. Chem. B 2009, 113, 10515–10520.
- Benko, Z.; Elder, R.T.; Liang, D.; Zhao, R.R. Fission yeast as a HTS platform for molecular probes of HIV-1 Vpr-induced cell death. Int. J. High Throughput Screen. 2010, 1, 151–162.
- Benko, Z.; Zhang, J.; Zhao, R.Y. Development of A Fission Yeast Cell-Based Platform for High Throughput Screening of HIV-1 Protease Inhibitors. Curr. HIV Res. 2019, 17, 429–440.
- Benko, Z.; Elder, R.T.; Li, G.; Liang, D.; Zhao, R.Y. HIV-1 Protease in the Fission Yeast Schizosaccharomyces pombe. PLoS ONE 2016, 11, e0151286.
- Benko, Z.; Liang, D.; Li, G.; Elder, R.T.; Sarkar, A.; Takayama, J.; Ghosh, A.K.; Zhao, R.Y. A fission yeast cell-based system for multidrug resistant HIV-1 proteases. Cell Biosci. 2017, 7, 5.
- Zhao, Y.; Elder, R.T. Yeast perspectives on HIV-1 Vpr. Front. Biosci. 2000, 5, 905–916.
- Andreola, M.L.; Litvak, S. Yeast and the AIDS virus: The odd couple. J. Biomed. Biotechnol. 2012, 2012, 549020.
- Chu, H.; Chan, J.F.; Yuen, T.T.; Shuai, H.; Yuan, S.; Wang, Y.; Hu, B.; Yip, C.C.; Tsang, J.O.; Huang, X.; et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: An observational study. Lancet Microbe 2020, 1, e14–e23.
