1. Introduction
1.1. General Introduction to Depression
Depression, an overarching term encompassing a spectrum of mood disturbances, stands as a predominant global health concern, currently recognized as a principal contributor to disability across the world. An estimated 5% of the global adult population is impacted by depression, a number witnessing an alarming ascent each year [
1,
2,
3]. Cumulative lifetime prevalence statistics, when evaluated on an international scale, suggest that about one in seven individuals will grapple with a depressive episode at some juncture in their lives, with rates ranging between 11.1% and 14.6% [
4]. Notably, while depressive episodes can manifest at various life stages, spanning early adolescence to adulthood, the median onset age gravitates around the mid-20s [
3,
5]. Gender disparities are also evident; women exhibit nearly double the susceptibility to experiencing depression during their lifetimes compared to their male counterparts, though this ratio demonstrates considerable cross-national variation [
6,
7].
The etiology of depression is multifaceted, interwoven with both environmental contributors such as socioeconomic conditions and biological determinants, including genetic predispositions [
3,
8]. The interplay between genes and the environment is further modulated by epigenetic mechanisms, adding another layer to its complexity [
9]. Among the various forms of depression, major depressive disorder (MDD) is particularly well-researched, although its pathophysiology is yet to be fully deciphered. Available evidence points towards molecular disruptions, including neurotransmitter imbalances and augmented inflammatory cytokine levels, underscoring their intricate molecular foundation [
10].
1.2. Current Therapeutic Approaches
The main goal of treatment for depression is an accurate diagnosis and the remission of symptoms. Depression is clinically identified as ‘major depression’ when it meets the symptomatic criteria outlined in the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, 2000 (DSM-V) [
11]. Diagnostic measures are based on clinical observation, although no objective biological markers currently exist. The main approaches to treatment can be separated into four broad categories: generic psychosocial interventions, formulation-based interventions of psychological therapy, pharmacotherapy, and electroconvulsive therapy (ECT) [
8]. In mild cases of depression, psychosocial and psychological intervention may be sufficient, while in more severe cases, pharmacotherapy is also required. Most pharmacological interventions for depression enhance the transmission of monoamines through different routes, with the most common first-line treatments being selective serotonin reuptake inhibitors and serotonin–norepinephrine reuptake inhibitors. Despite the range of psychological and pharmacological therapies available, many patients do not achieve remission [
12,
13].
1.3. General Introduction to miRNAs
MicroRNAs are small non-coding RNAs of approximately 19–22 nucleotides that function to inhibit the level of target genes within discrete regulatory networks [
14]. Over 60% of human protein-coding genes contain conserved miRNA target sites [
15]. The interaction networks of miRNAs are complex and vast; one miRNA can target many mRNAs, and one mRNA can be regulated by many different miRNAs [
16,
17]. Their identification in many different organisms also suggests their regulatory mechanisms are evolutionarily conserved [
18]. According to recent work by the Functional Annotation of the Mammalian Genome (FANTOM5) consortium, approximately half of miRNAs show cell type specificity, and one-quarter are more broadly expressed [
19].
In the central nervous system, miRNAs play a role in neurodevelopment, synaptic plasticity, and stress responses, thus providing a molecular link to psychiatric conditions [
20,
21,
22]. Several lines of evidence have already shown their involvement in other neurological and mental health disorders [
22].
In fact, miRNAs are differentially expressed in depression, as identified by a multitude of recent studies, pointing to their involvement here as well [
23]. miRNAs have emerged as key players in depression due to their role as post-transcriptional regulators of gene expression and their ability to pass the blood–brain barrier [
24]. Excitingly, they offer new avenues for understanding the intricate molecular underpinnings of depression. Their capacity to modulate multiple gene networks associated with neuroplasticity and stress response, as well as their shuttling between the blood and the brain, makes miRNAs compelling targets for both diagnostic and therapeutic interventions in depressive disorders.
2. MicroRNAs and Depression
2.1. miRNAs in the Pathophysiology of Depression
Emerging research underscores noticeable shifts in miRNA levels in both the brain and periphery of individuals with depression. Generally, the predominant research paradigms encompass the following: (a) a comparative study of miRNA profiles between patients and controls; (b) an exploration of miRNA modulations subsequent to antidepressant interventions. In the former approach, the prefrontal cortex emerges as a primary brain region of interest. In the periphery, researchers often rely on whole blood, plasma, or serum samples. The second methodology is predominantly engaged to discern miRNA shifts following therapeutic regimens. A less conventional, albeit more invasive, medium is cerebrospinal fluid to probe miRNA dysregulation. Downstream analysis usually involves bioinformatics-driven approaches to identify miRNA–mRNA interactions, facilitating an understanding of the broader regulatory networks at play. Furthermore, to decode the functional dynamics of miRNA regulation, in vitro evaluations and animal model studies are occasionally integrated, though they are less prevalent.
Some of the first studies to identify the role of microRNAs in the pathophysiology of MDD have examined the postmortem human brain. In one example study, the levels of miR-182 expression were disrupted in dentate gyrus granule cells of individuals with depression [
76,
77]. In the anterior cingulate cortex, a brain region critical for emotional and mood regulation, a depression-specific miRNA expression profile was identified compared to controls [
78]. In a similar approach, miR-1202 was found to be downregulated in the prefrontal cortex of individuals with depression compared to controls [
79]. This microRNA was found to be enriched in the brain ad negatively correlated with
GRM4 expression, a metabotropic glutamate receptor. Importantly, this study also showed that in individuals who had attained remission, levels of miR-1202 were increased.
While the postmortem brain remains a key system for investigating the molecular mechanisms of MDD, a drawback remains that microRNAome profiling in brain tissue at the time of death or sample collection may not be reflective of the disease course [
22]. In recent years, miRNAs have been assessed in the periphery of individuals with depression. An advantage of this approach is capturing real-time miRNA differences between patient and control populations and quantifying miRNA changes in patients in response to treatment. One of the first studies to take this approach showed decreased miR-135a levels in patients with depression compared to controls, which had increased after 3 months of selective serotonin reuptake inhibitor treatment or cognitive behavioral therapy [
80]. Another study by Belzeaux and colleagues identified a distinct set of miRNAs that are dysregulated in patients with depression, which could predict clinical improvement and treatment response [
81]. A further study showed that with electroconvulsive therapy for treatment-resistant depression, levels of miR-223-3p could predict treatment response [
82].
Despite numerous studies, there remains a substantial lack of consensus on key miRNAs and their functions in depression. To address this, recent advancements in the field have developed methods for the isolation of cell-type-specific microRNAs in the periphery. For example, microRNAs originating in different brain cell types can now be selected. Pioneering work taking this approach has examined miRNAs within neuron-derived extracellular vesicles in the periphery and has found decreased miR-93 levels in individuals with depression [
83]. In another study comparing the neuron-derived extracellular vesicle microRNAome before and after treatment with escitalopram, a signature of miR-21-5p, miR-30d-5p, and miR-486-5p was found to be associated with treatment response [
84].
2.2. miRNAs as Diagnostic Biomarkers
The search for non-invasive peripheral biomarkers that mirror changes in the brain is a high priority in research on depression. Such biomarkers could substantially impact current clinical diagnosis practices and, if capable of predicting antidepressant response, the assignment of efficacious treatments. In this regard, circulating miRNAs in blood and other bodily fluids have emerged as promising candidates. Owing to their remarkable stability in circulation and the detectability of brain-specific miRNAs such as miR-9 and miR-223-3p [
82,
88], miRNAs offer a new avenue in biomarker research.
2.2.1. Blood-Derived miRNAs: Refining the Complex Landscape of Depression Biomarkers
Differential expression of miRNAs in the periphery has been consistently identified in depression and following treatment with antidepressants. Thus, it is clear that the etiology and therapeutic intervention in depression rely at least in part on regulation by miRNAs.
One seminal study highlighted the plasticity of miRNA profiles following treatment, showing that a twelve-week therapeutic regimen of escitalopram promoted the upregulation of 28 microRNAs while concomitantly downregulating miR-34c-5p and miR-770-5p in the blood of individuals with depression [
87,
89] (see
Table 1). In another study, an eight-week citalopram treatment normalized blood concentrations of two deregulated miRNAs, miRNA-132 and miR-124 [
90]. While a four-week intervention with citalopram normalized the levels of 10 previously deregulated miRNAs [
91]. A particularly noteworthy observation common to these therapeutic interventions was the consistent upregulation of miR-335 [
89,
91], previously shown to regulate glutamate metabotropic receptor 4 (
GRM4), implicated with anxiety-associated behaviors [
91]. Another study observed that blood levels of miR-1202 decreased in individuals with depression, a trend that was rectified following an eight-week treatment course of either escitalopram or desvenlafaxine [
79,
92]. Notably, both miR-1202 and miR-335 have been implicated in regulating GRM4 signaling pathways, thereby influencing stress-related behaviors [
79]. Further adding to the complexity, a different study indicated that treatment responses to escitalopram, desvenlafaxine, and duloxetine were associated with changes in miR-1202 and miR-16-5p levels [
93].
Table 1. Comparative studies on peripheral miRNA expression in depression: pre- and post-antidepressant treatment effects.
| Refs. |
Tissue Source |
miRNAs |
Status (Patient vs. Control) |
Antidepressant Treatment |
Regulation by Treatment |
miRNAs |
| [79,92] |
Whole blood |
miR-1202 |
Down |
Citalopram |
Up |
miR-1202 |
| [87,89] |
Whole blood |
- |
- |
Escitalopram
(12 weeks) |
Up |
miR-130b, miR-505, miR-29b-2, miR-26b, miR-22, miR-26a, miR-664, miR-494, let-7d, let-7g, let-7e, let-7f, miR-629, miR-106b, miR-103, miR-191, miR-128, miR-502-3p, miR-374b, miR-132, miR-30d, miR-500, miR-589, miR-183, miR-574-3p, miR-140-3p, miR-335, miR-361-5p |
| Down |
miR-34c-5p, miR-770-5p |
| [90] |
Whole blood |
miR-132-3p, miR-124-3p |
Up |
Citalopram
(8 weeks) |
Up |
miR-124 |
| Down |
miR-132-3p |
| [94] |
Whole blood |
- |
- |
Duloxetine
(8 weeks) |
Up |
miR-3688, miR-5695 |
| [95] |
Whole blood |
- |
- |
Escitalopram
(2 weeks) |
Up |
miR-103a-3p, miR-103b, miR-106a-5p, miR-106b-3p, miR-140-3p, miR-145-5p, miR-148b-3p, miR-151a-5p, miR-15a-5p, miR-15b-5p, miR-17-5p, miR-182-5p, miR-185-3p, miR-185-5p, miR-186-5p, miR-20a-5p, miR-20b-5p, miR-210-3p, miR-25-3p, miR-30a-5p, miR-30b-5p, miR-3158-3p, miR-3158-5p, miR-324-5p, miR-331-5p, miR-500a-3p, miR-502-3p, miR-532-5p, miR-550a-3p, miR-584-5p, miR-589-5p, miR-660-5p, miR-93-5p |
| Down |
miR-1301-3p, miR-191-3p, miR-200b-3p, miR-222-3p, miR-25-5p, miR-27a-3p, miR-30c-1-3p, miR-3168, miR-328-3p, miR-505-5p, miR-744-5p, miR-92a-1-5p |
| [96] |
Plasma |
miR-144-5p |
Down |
Personalized
(8 weeks) |
Up |
miR-144-5p |
| [97] |
Serum |
miRNA-34a-5p, miRNA-221-3p |
Up |
Paroxetine
(8 weeks) |
Down |
miRNA-34a-5p, miRNA-221-3p |
| miRNA-451a |
Down |
Up |
miRNA-451a |
| [84] |
Neuron-derived EV |
- |
- |
Escitalopram
(8 weeks) |
Up
(responders) |
miR-30d-5p and miR-486-5p |
In parallel to the analysis of discrete microRNAs, several studies have opted for a more comprehensive approach employing small RNA sequencing (sRNA-seq). Using this approach, Belzeaux et al. showed that miR-3688 and miR-5695 served as predictive markers for exacerbated suicidal ideation in individuals subjected to an eight-week course of duloxetine or a placebo [
94]. Similarly, Yrondi et al. employed next-generation sequencing (NGS) and observed that pharmacological interventions with escitalopram affected levels of 45 microRNAs, with miR-185-5p being particularly correlated with the reduced incidence of nausea, a common side effect associated with selective serotonin reuptake inhibitors (SSRIs) [
95]. In another study, Lopez et al. utilized small RNA sequencing to investigate miRNAs as potential biomarkers for antidepressant responses in MDD within a randomized placebo-controlled trial of duloxetine. The research, conducted before and 8 weeks post-treatment, identified that miR-146a-5p, miR-146b-5p, miR-425-3p, and miR-24-3p exhibited differential expression linked to treatment outcomes. Furthermore, these miRNAs were shown to regulate the MAPK/Wnt signaling pathways, suggesting their significant role in the antidepressant response [
86].
In sum, these studies substantiate the proposed use of circulating miRNAs in blood as biomarkers for depressive disorders and treatment responses. Moreover, the findings highlight the complex molecular interplay underlying depression and pharmacological interventions, setting the stage for future research that can enable more targeted and efficacious therapeutic strategies.
2.2.2. Fluid-Based miRNA Landscapes: Serum, Plasma, and Cerebrospinal Fluid
The body of evidence linking specific miRNA profiles to depression extends beyond blood to encompass serum, plasma, and cerebrospinal fluid (CSF). For instance, two studies reported elevated serum and plasma levels of miR-182 and miR-132 in MDD, with both miRNAs found to modulate BDNF expression in neuronal cells [
77,
90]. Key studies show that inhibiting miR-182-5p alleviates depression-like behaviors in mouse models, suggesting its potential as a therapeutic target [
98]. The upregulation of miR-182 reduces BDNF in the hippocampus, further underlining its role in depression pathophysiology [
85]. A separate investigation by Wang et al. revealed that diminished plasma levels of miR-144-5p were correlated with depressive symptoms and reversed following eight weeks of personalized antidepressant treatment [
96]. Rodent models also imply a role for miR-144-5p in hippocampal functioning, specifically through the regulation of the PTP1B protein, a known modulator of BDNF/TrkB signaling [
99]. Several studies reported that many miRNAs, including miR-1202, miR-135a-5p, miR-184, let-7g-5p, miR-103a-3p, miR-107, miR-16-5p, miR-34a-5p, miR-451a, miR-221-3p, and let-7d-3p, found in CSF and serum, were dysregulated in depression and responded to antidepressant treatments [
97,
100,
101,
102,
103].
2.2.3. Extracellular Vesicles: Emerging Protagonists in the Biomarker Landscape of Depression
In the exploration of biomarkers for depression, extracellular vesicles (EVs) have recently come under investigation [
104]. As previously described, EVs serve as a robust reservoir of miRNAs, facilitating their circulation between the brain and peripheral blood, and vice versa. EVs have been implicated in inflammatory disorders [
105,
106], cancer [
107], and neurodegenerative diseases [
108], with their role in psychiatric conditions gaining increasing interest [
109]. For example, high-throughput miRNA sequencing identified differentially expressed miRNAs in the serum exosomes of patients with depressive symptoms [
24]. Upregulated miRNAs were linked to the modulation of neurotrophic signaling pathways, while downregulated ones were associated with the regulation of apoptosis and immune functions [
24]. Notably, the levels of miR-139-5p in serum exosomes distinguished individuals with MDD from healthy controls and could also induce depressive-like behaviors in murine models [
110].
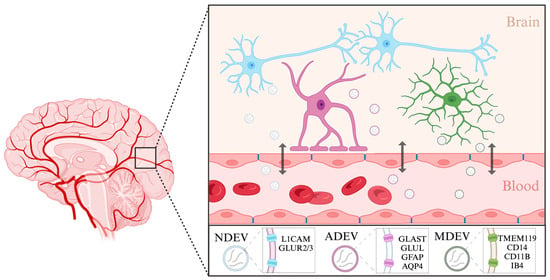
Of particular interest is the potential for isolating brain-derived extracellular vesicles (BDEVs) from peripheral blood, which offers a window into central nervous system-specific molecular landscapes [
111,
112]. BDEVs can originate from many different cell types in the brain, including neurons, astrocytes, and microglia (
Figure 1). Neuron-derived EVs have been isolated previously using a biotinylated antibody to L1 cell adhesion molecule (L1CAM), a transmembrane protein specific to neurons [
113]. They could also be labeled using antibodies against the GluR2/3 subunits of AMPA receptors [
114]. Using this approach, studies have already begun to assess the roles of neuron-derived EVs in depression. Elevated levels of miR-17-5p in neuron-derived EVs correlated with subthreshold depressive symptoms [
112]. Moreover, neuronal EVs isolated from individuals with depression undergoing 8-week escitalopram treatment displayed brain-specific proteins and miRNAs (such as miR-30d-5p and miR-486-5p) and also exhibited size alterations reversible by the antidepressant therapy. Changes in miRNA cargo in these neuron-derived EVs also reflected varying responses to antidepressant interventions [
84].
Figure 1. Characterization of brain-derived extracellular vesicles. Brain-derived extracellular vesicles enable cell-to-cell communication between different cell types in the brain as well as in the periphery. Extracellular vesicles can originate in neurons (NDEVs), astrocytes (ADEVs), and microglia (MDEVs). These vesicles can shuttle across the blood–brain barrier and enter the body’s circulation. Listed are the markers specific to each cell type that have been used to isolate extracellular vesicles from solution. Created with BioRender.com. (Retrieved from
https://app.biorender.com/biorender-templates, accessed on 30 January 2024).
The isolation of astrocyte and microglia-derived EVs is also starting to be explored. One of the main forms of communication between neurons and astrocytes is through EVs, where they can have both neuroprotective and pathological properties, highlighting their importance [
115]. Astrocyte-derived EVs (ADEVs) have previously been isolated from human blood using a biotinylated antibody to glutamine aspartate transporter (GLAST), an astrocyte-specific membrane protein, glutamine synthetase (GLUL) [
116], as well as using glial fibrillary acidic protein (GFAP) and aquaporin 4 (AQP4) [
117]. Using this approach, ADEVs have been implicated in modulating immune responses during stress and depression [
118,
119]. It has been shown that individuals with depression have larger numbers of GFAP and AQP4/GFAP-positive EVs in the blood than healthy controls, suggesting increased leakage of astrocyte-derived EVs through the blood–brain barrier. In a recent study, the miRNA content of ADEVs from individuals with depression was found to have significantly deregulated miR-9 levels, suggesting a specific involvement of this miRNA in the disease’s pathophysiology [
120].
Similar to astrocytes, microglia also rely on EVs for cell-to-cell communication within the brain. Largely, the isolation of microglia-derived EVs from complex fluids such as blood samples has so far also been conducted using antibodies or lectins against microglial surface markers [
121]. The heterogenous nature of microglial cells is inevitably reflected in their surface protein content and cargo and should thus be used to inform isolation protocols. For instance, in a stroke experimental model, EVs from microglia were isolated using markers characteristic of their specific activation state, namely TMEM119 and CD14 [
122]. In another example, small EVs of microglial origin were isolated using CD11B, a more general marker of microglia [
123]. Microglia or macrophage-derived EVs have also been isolated using myeloid marker IB4 [
124]. Examining the contents of cell-type specific EVs enables a more targeted understanding of disease mechanisms. Further efforts to characterize extracellular vesicles and their components which originate in different brain cell types will improve our understanding of disease mechanisms.
This entry is adapted from the peer-reviewed paper 10.3390/ijms25052866