Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Serum albumin is a popular macromolecule for studying the effect of proteins on the colloidal stability of nanoparticle (NP) dispersions, as well as the protein–nanoparticle interaction and protein corona formation.
- serum albumin
- shungite carbon nanoparticles
- fatty acids
1. Serum Albumin and Shungite Carbon Nanoparticles
In recent years, nanoparticles have been widely used in practical biomedicine [1]. This is due to their high penetrating ability into tissues and cells, the ability to transport biologically active substances, their redox properties, and their effects on the structural organization of biosystems [2]. Simultaneously, more and more data are accumulating on the potential risk of the long-term use of nanoparticles [3] and their toxicity to the environment [4]. Inorganic nanoparticles based on silicon oxide, various metals, their oxides, and sulfides, as well as quantum dots and carbon nanoparticles, attract special attention. However, insufficient knowledge of the possible solvent-dependent physicochemical mechanisms of the interaction of nanoparticles with biological objects and with protein systems, in particular, leads to the absence of theoretical models that make it possible to predict the result of nanoparticle interactions with biosystems [5].
Carbon nanomaterials present the best form of abiogenic nanomaterials found in the environment. In particular, natural shungite carbon (ShC) is widely used in cosmetics and foods without even passing full safety tests [6]. It is actively used for various cosmetic and balneological purposes, as bioadditives, and filters for water treatment. However, such an application has a purely marketing value without sufficient scientific justification. In spite of a firmly justified molecule-based structure of the solid [7], the molecular mechanisms of its biological activity have not been characterized enough. At the same time, the development of green technologies [8] for obtaining relatively large amounts of ShC nanoparticles (ShC NPs), which are characterized by structural homogeneity [9], makes ShC a promising material for bionanomedicine purposes.
Shungites are widespread in Karelia (Russia) as rocks of the Precambrian age [10][11]. They generally present a mixture of nanosized crystalline silica with sp2 carbon, the mass content of which varies from 5% to 98%, depending on the deposit. Shungite rocks originated in soft geological conditions (temperature less than 450 °C, pressure 70 MPa), and the water environment played a key role in their formation process. ShC can be extracted from carbon-rich shungite rocks based on green chemistry methods [10] with the formation of stable dispersions of nanoparticles (ShC NPs) in the aquatic environment. A possible scenario for the origin of the ShC was proposed [12] based on the hydrothermal impact in the condition of an oxygen anomaly on the graphite layers that existed in the region in the Precambrian period. Subsequently, oxidized graphenes were subjected to reduction processes over 1.8 billion years in a water environment. At the same time, the presence of residual and constitutional water in the nanocarbon structure makes it easy to release ShC NPs into the aquatic environment, thereby ensuring the unique properties of ShC to re-disperse when water is added.
ShC NPs are formed by basic structural units (BSUs) less than 1 nm, which are necklaced graphene molecules. This reflects the idea that graphene molecules are always terminated by heteroatoms, which form a kind of “necklace” on the graphene domains [7]. It was shown empirically and virtually that the BSUs belong to a large family of reduced graphene oxides (rGOs). In a liquid environment, they retain their characteristic tendency to form nanosized stacks, thus self-associating and forming nanoparticles [10][12]. The morphology and spectral properties of the obtained ShC dispersions depend on the liquid in use. The most stable dispersions with NP concentrations of ca 0.1 mg/mL are formed in water, although the dispersions in toluene and other solvents [13] can be produced as well.
Serum albumin (SA) has a broad perspective in the creation of bionanoconjugates with nanoparticles for the regulation of the protein assembly into fibrils and the disaggregation of preformed fibrils [14], as well as being an ideal candidate for nanoparticle formulation as potential controlled-release drug delivery systems [15]. In addition, SA has been used for surface coating and passivation in biological liquids [16][17] and as biosurfactants [18]. In turn, SA is used as a sensor of conformational changes when interacting with surfaces of various natures [19]. Such a range of possible applications of albumin is due to both the availability and cheapness of SAs and the relative completeness of knowledge [16][20][21] of their conformational state, intermolecular interaction, colloidal stability [22], and phase behavior in dispersions with different microenvironments.
When the biological environment interacts with the surface, colloidal nanomaterials, nanoparticles, a dynamic coating of biomolecules, and proteins spontaneously arise around them, called the protein corona [19][23][24] (Figure 1). Irreversible or at least long-term binding of a protein by a nanoparticle leads to the formation of a “hard corona”, whereas the rapidly reversible binding of a protein that has a higher turnover rate results in the formation of a “soft corona”. The prospects for the wide use of nanoparticles in biomedicine [1][3] suggest the need to study this phenomenon since the toxicity of nanomaterials and the nanorisks of their use largely depend on the functioning of the corona. Blood plasma proteins, and in particular albumins, are the most important components in the formation of the protein corona, although the mechanism of its formation is complex and insufficiently studied [25], being a phase-regulating process.
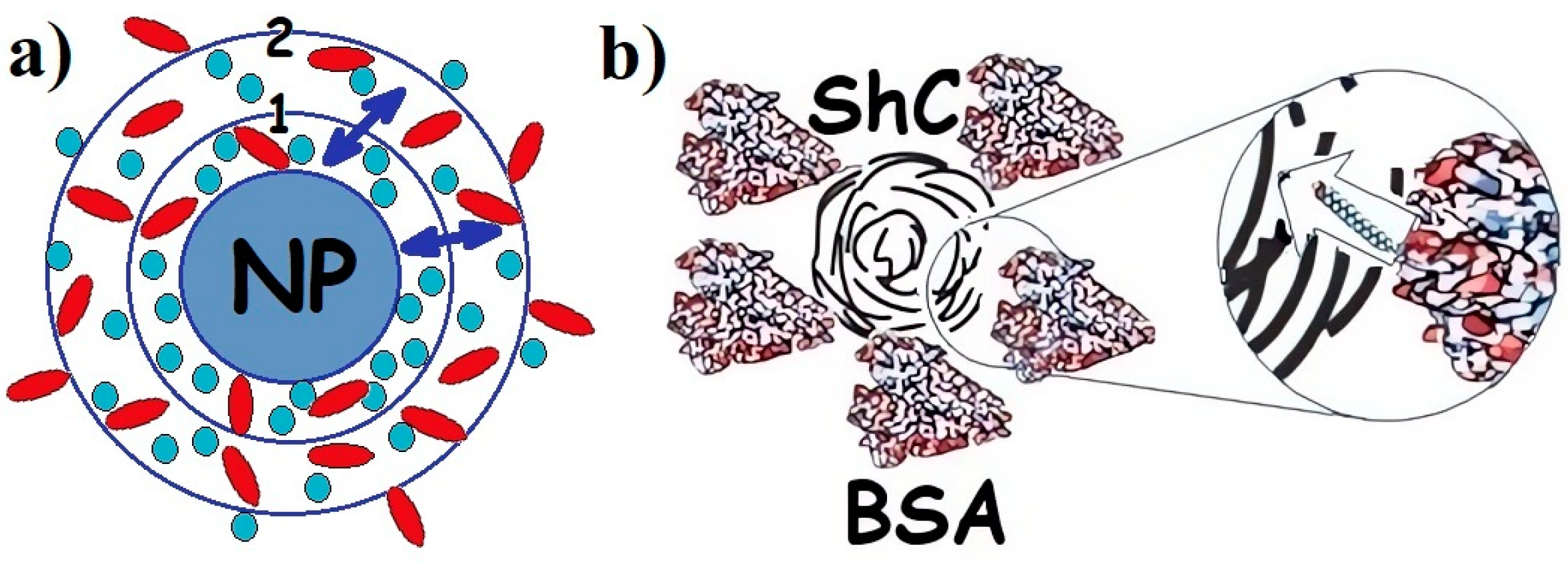
Figure 1. (a) Schematic presentation of protein corona in the dispersion of any nanoparticle (NPs) with proteins of different types: 1—hard corona; 2—soft corona; (↔)—exchange of proteins between hard and soft corona. Proteins are divided between hard and soft corona depending on the affinity of the proteins for the NP surface. (b). Corona of bovine serum albumin (BSA) around shungite carbon NP. Insert: long rod indicates exchange of fatty acids (which are not a part of the corona) between BSA and NP. Different colors are provided for ease of observation.
Highly organized protein molecules are edited by evolution to perform specific functions. However, other supramolecular structures and nanoparticles have been intensively introduced into living systems for possible adjustment of these functions. It is natural to assume that, in this case, the introduced structures, to some extent, must also fulfill a certain norm of function per unit of structure, characteristic of biomacromolecules. In this regard, the question arises whether the nanoparticles themselves are capable of performing similar functions characteristic of albumin. It can be assumed that in the case of such abilities, hybrid dispersions of albumin and nanoparticles will be the most productive in biomedical applications due to the possibility of mutually complementing and regulating functions. In this regard, shungite carbon in the form of ShC NPs [10] is of particular interest.
The results obtained provide information that the incorporation of molecules into defects in the water hydrogen bond system allows this water system to control the incorporation processes either by changing macromolecular conformation or by the emergence of supramolecular or cluster organization in the newly created defects. The formation of a multilevel heterogeneous cluster system of ShC NPs is a thermodynamic process (although it is accompanied by kinetic phenomena).
2. Structure-Function Properties of SA
Serum albumin (SA) is a transport protein in the blood. Its content is 55–60% of the total amount of proteins in plasma, and the concentration is about 30–50 mg/mL [20]. The ability of albumin to bind chemical compounds of various structures: hormones, metabolic products, toxins, drugs, metal ions, and fatty acids (FAs), as well as to maintain a sufficiently high osmotic pressure in plasma, comparable to the pressure in the cytoplasm, determines the variety of albumin functions in the vital activity of organisms [20]. The serum albumin is a poly-functional communicative and integrative molecular system that performs a homeostatic function by regulating both inter-tissue and inter-organ processes, and the functional and metabolic activity of inter-tissue processes [26]. SA has repeatedly served as a model protein for studying the osmotic properties of biopolymer solutions and the mechanisms that cause the behavior of protein solutions to deviate from ideal [27][28][29]. Based on its physicochemical parameters, various models have been constructed for the theoretical and experimental analysis of protein–protein intermolecular interactions [30]. Thus, SA solution is a completely self-sufficient system for studying the molecular mechanisms of homeostatic reactions in the presence of metabolites, drags, and nanoparticles of various natures. Basic details of the molecular structure of albumin are shown in Figure 2.
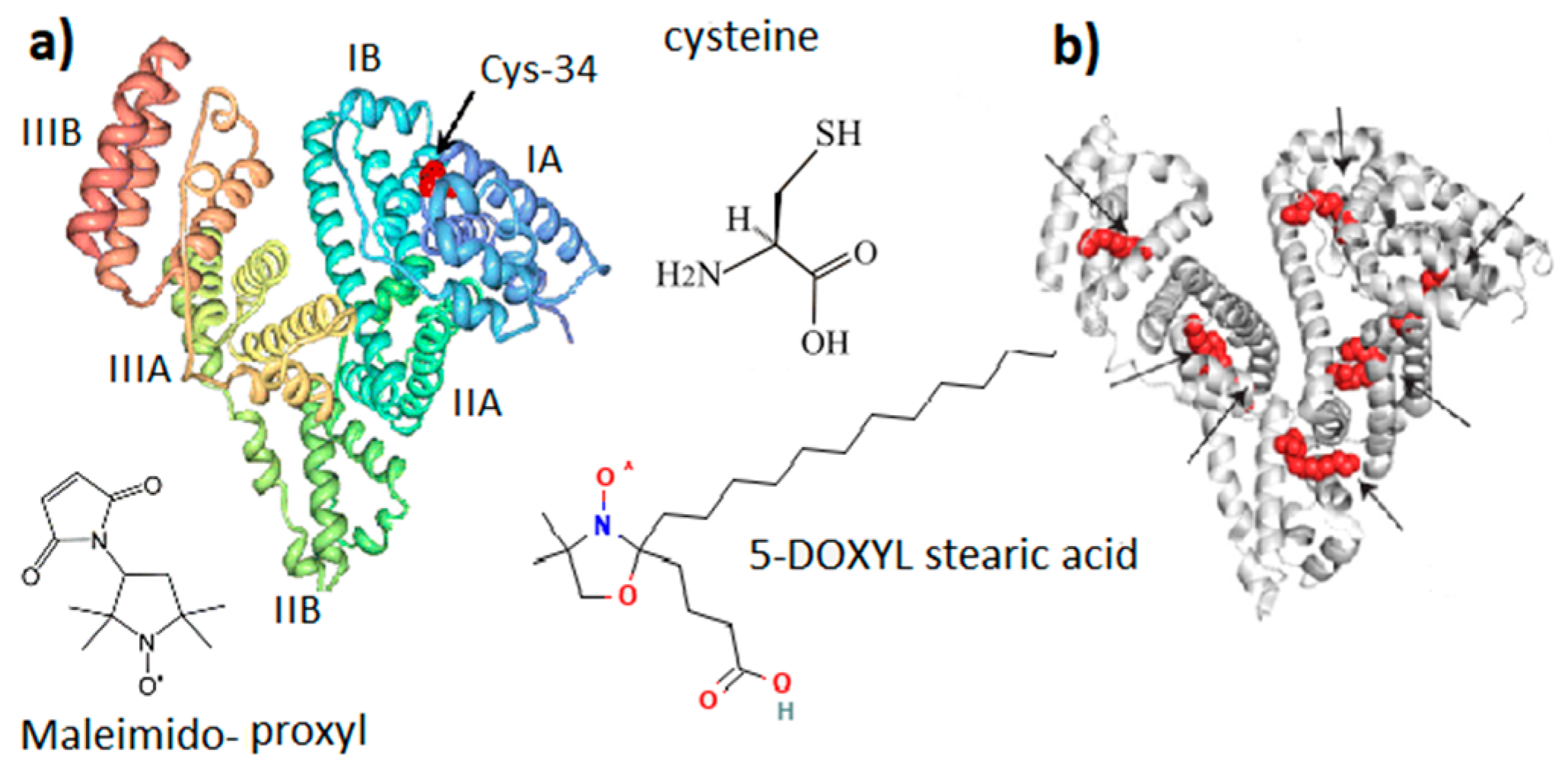
Figure 2. General molecular structure of serum albumin (SA). (a) SA domain structure with Cys-34 located in the IA domain cavity. (b) Potential binding sites of fatty acid in the SA structure. Maleimido-proxyl is a spin label that modifies the SH group of Cys-34. 5-DOXYL stearic acid is a spin probe adsorbed at fatty acid binding sites.
Water albumin solutions were found to contain significant fractions of dimers and high-order oligomers. Human SA is the most capable of oligomerization. In addition, the protein tends to form thixotropic structures, the structural elements of which are protein associates—clusters [31][32]. Cluster formation is probably a thermodynamic process of phase transition described by phase diagrams and relevant to protein conformation [33][34][35]. The structural heterogeneity of SA, caused by both genetic and postsynthetic factors, leaves many questions regarding the relationship between heterogeneity and functional properties, as well as the mechanism of “unloading” from ligands [36].
The albumin heterogeneity does not make it possible to obtain high-quality protein crystals suitable for a high-resolution X-ray diffraction analysis. Nevertheless, great efforts to obtain more than half a dozen different crystalline forms of SA contributed to the identification of the X-ray structure of SA with a resolution of 0.25 nm [37]. It was confirmed [38] that the three-dimensional structure of SA with dimensions of 14 nm in length and 4 nm in diameter is organized into three main domains (I, II, III), each containing two subdomains (A and B). Heart-shaped subdomains are built from 3–4 α-helices. The structures of the main domains coincide with mutual superpositions. Subdomains IA, IB, and IIA are densely grouped and form a “head”, while subdomains IIB, IIIA, and IIIB form an elongated “tail”.
Domain I of SA is believed to be responsible mainly for the binding of metal ions [39]. Domain II and III both have pockets formed by hydrophobic and positively charged residues in which different compounds, such as bilirubin, pyridoxal, and partially fatty acids, can be accommodated. Three tentative binding sites for FA, each with different surroundings, are located at the surface of each domain. In the presence of FAs, SA is an equilibrium mixture of macromolecules with different degrees of binding site occupation, which are in different conformations. The binding of FAs changes the charge equilibrium in the protein and, as a result, changes the interdomain interactions and the conformation of the protein [40]. As a result, different fractions of the protein can appear.
Along with maintaining colloid–osmotic tissue homeostasis and binding and transporting physiologically important molecules and ions, SA has an important antioxidant function due to the ability of the SH group of the amino acid cysteine (Figure 2) to interact with free radicals and reactive oxygen species [26][41]. Human SA (HSA) is a single polypeptide chain with 585 residues containing 35 cysteines, which form 17 disulfide bridges, leaving the SH group of cysteine in the 34 position of the polypeptide chain (Cys-34, Figure 2) free. It is localized in a hydrophobic cavity about 1 nm deep in domain I [42]. The binding of one and two fatty acids (5-DOXYL stearic acid in Figure 2) significantly affects both the opening of the Cys-34 cavity and the global unfolding of the albumin structure, which increases the mobility of the atomic groups that form the Cys-34 cavity. In terms of structure and function, HSA and bovine SA (BSA) are very similar [43]. BSA is formed from 583 amino acid residues, 76% of which are identical to HSA. On the other hand, a number of differences in the binding pockets, surface structures, and charge distribution were found. Since HSA contains one tryptophan, this protein is more convenient for optical fluorescence studies. However, for studying the interaction of a protein with other molecules and nanoparticles, BSA is preferable because it is less hydrophobic and less prone to aggregation and self-association, forming particles with smaller sizes and narrower polydispersity.
The structural-dynamic state of albumin molecules is characterized by a number of reversible conformational transitions in the region of non-physiological pH [28][43]. At neutral pH, SA exists in the neutral (N) form. At basic pH, the N form shifts to the basic (B) form (at pH 8) and to the “aged” (A) form at pH 10. At low pH (4.3), SA undergoes a transition to the fast (F) form and the extended (E) form at pH 2.7. For both acidic and basic transitions, SA undergoes an unfolding and loosening of its structure (see Figure 2 presented in ref. [28]).
Increasing the temperature to 55 °C causes a gradual and reversible change in the protein secondary structure. The most significant change includes more content of helix secondary structure (about 3%) at 40 °C relative to that at 45°. It is accompanied by the reversible separating of I and II domains around 42–43 °C [44][45]. The region of 58–65 °C is characterized by a sharp onset of irreversible denaturation processes accompanied by protein aggregation [46].
FA binding under physiological conditions also causes local and global changes, manifested both at the level of the relative mutual displacement of I, II, and III domains of the SA structure and in smaller-scale changes in the region of the SH group of the Cys-34 [41]. This SH group is believed to be in two microstates: the reduced form, predominantly in the closed cavity, and the oxidized form when the cavity opens. The nearby amino acid residues histidine (His-39) and tyrosine (Tyr-84) located in the IA domain also affect the reactivity of the SH- group of Cys-34 [41]. Interactions with FAs not in direct contact with Cys-34 also induce conformational changes in its environment that promote the formation of -S-S-disulfide [47]. Albumin oxidation at other amino acid residues also affects, to some extent, protein thermal stability, resistance to aggregation, and aggregate morphology [48].
This entry is adapted from the peer-reviewed paper 10.3390/ijms25052465
References
- Nguyen, V.H.; Lee, B.J. Protein corona: A new approach for nanomedicine design. Int. J. Nanomed. 2017, 12, 3137–3151.
- Li, D.; Zhang, W.; Yu, X.; Wang, Z.; Su, Z.; Wei, G. When biomolecules meet graphene: From molecule-level interactions to material design and applications. Nanoscale 2016, 8, 19491–19509.
- Wang, Y.; Cai, R.; Chen, C. The Nano-Bio Interactions of Nanomedicines: Understanding the Biochemical Driving Forces and Redox Reactions. Acc. Chem. Res. 2019, 52, 1507–1518.
- Nasser, F.; Constantinou, J.; Lynch, I. Nanomaterials in the Environment Acquire an “Eco-Corona” Impacting their Toxicity to Daphnia Magna—a Call for Updating Toxicity Testing Policies. Proteomics 2020, 20, 1800412.
- Yang, W.; Wang, L.; Mettenbrink, E.M.; De Angelis, P.L.; Wilhelm, S. Nanoparticle toxicology. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 269–289.
- Abaeva, L.F.; Shumsky, V.I.; Petritskaya, E.N.; Rogatkin, D.A.; Lubchenko, P.N. Nanoparticles and nanotechnologies today and beyond. Alm. Clin. Med. 2010, 22, 10–16. (In Russian)
- Sheka, E. A Neoteric View of sp2 Amorphous Carbon. Nanomaterials 2023, 13, 1648.
- Rozhkova, N.N.; Gorlenko, L.E.; Emel’yanova, G.I.; Korobov, M.V.; Lunin, V.V.; Ôsawa, E. The effect of ozone on the structure and physico-chemical properties of ultradisperse diamond and shungite nanocarbon elements. Pure Appl. Chem. 2009, 81, 2093–2105.
- Rozhkova, N.N. Aggregation and stabilization shungite carbon nanoparticles. Russ. J. Gen. Chem. 2013, 83, 2676–2685.
- Rozhkova, N.N.; Rozhkov, S.P.; Goryunov, A.S. Natural Graphene-Based Shungite Nanocarbon. In Carbon Nanomaterials Sourcebook: Graphene, Fullerenes, Nanotubes, and Nanodiamonds; Aliofkhazraei, M., Ali, N., Milne, W.I., Eds.; CRC Press Inc. (Taylor and Francis Group): Boka Raton, FL, USA; London, UK; New York, NY, USA, 2016; Volume 1, pp. 153–178.
- Buseck, P.R.; Galdobina, L.P.; Kovalevskii, V.V.; Rozhkova, N.N.; Valley, J.W.; Zaidenberg, A.Z. Shungites: The C-rich rocks of Karelia, Russia. Can. Mineral. 1997, 35, 1363–1378.
- Sheka, E.F.; Rozhkova, N.N. Shungite as the natural pantry of nanoscale reduced graphene oxide. Int. J. Smart Nano Mater. 2014, 5, 1–16.
- Razbirin, B.S.; Rozhkova, N.N.; Sheka, E.F.; Nelson, D.K.; Starukhin, A.N. Fractals of graphene quantum dots in photoluminescence of shungite. JETP 2014, 118, 735–746.
- Zaman, M.; Ahmad, E.; Qadeer, A. Nanoparticles in relation to peptide and protein aggregation. Int. J. Nanomed. 2014, 9, 899–912.
- Elzoghby, A.O.; Samy, W.M.; Elgindy, N.A. Albumin-based nanoparticles as potential controlled release drug delivery systems. J. Control Release 2012, 157, 168–182.
- Park, J.H.; Jackman, J.A.; Ferhan, A.R.; Ma, G.J.; Yoon, B.K.; Cho, N.J. Temperature-Induced Denaturation of BSA Protein Molecules for Improved Surface Passivation Coatings. ACS Appl. Mater. Interfaces. 2018, 10, 32047–32057.
- Schuberta, J.; Chanana, M. Coating Matters: Review on Colloidal Stability of Nanoparticles with Biocompatible Coatings in Biological Media, Living Cells and Organisms. Curr. Med. Chem. 2018, 25, 4553–4586.
- Otzen, D.E. Proteins in a brave new surfactant world. Curr. Opin. Colloid Interface Sci. 2015, 20, 161–169.
- Treuel, L.; Brandholt, S.; Maffre, P. Impact of Protein Modification on the Protein Corona on Nanoparticles and nanoparticle-Cell Interactions. ACS Nano 2014, 8, 503–513.
- Peters, T., Jr. All about Albumin: Biochemistry, Genetics and Medical Applications; Academic Press, Inc.: San Diego, CA, USA, 1996; p. 432.
- Rosenoer, V.M.; Oratz, M.; Rothschild, M.A. Albumin: Structure, Function and Uses; Elsevier: Amsterdam, The Netherlands, 2014; p. 412.
- Sun, B.; Zhang, Y.; Chen, W.; Wang, K.; Zhu, L. Concentration Dependent Effects of Bovine Serum Albumin on Graphene Oxide Colloidal Stability in Aquatic Environment. Environ. Sci. Technol. 2018, 52, 7212–7219.
- Lundqvist, M.; Stigler, J.; Elia, G.; Lynch, I.; Cedervall, T.; Dawson, K.A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. USA 2008, 105, 14265–14270.
- Monopoli, M.P.; Walczyk, D.; Campbell, A.; Elia, G.; Lynch, I.; Baldelli Bombelli, F.; Dawson, K.A. Physical-Chemical Aspects of Protein Corona: Relevance to In Vitro and In Vivo Biological Impacts of Nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534.
- Kopac, T. Protein corona, understanding the nanoparticle-protein interactions and future perspectives: A critical review. Int. J. Biol. Macromol. 2021, 169, 290–301.
- Mishra, V.; Heath, R.J. Structural and Biochemical Features of Human Serum Albumin Essential for Eukaryotic Cell Culture. Int. J. Mol. Sci. 2021, 22, 8411.
- Vilker, V.L.; Colton, C.K.; Smith, K.A. The osmotic pressure of concentrated protein solutions: Effect of concentration and pH in saline solutions of bovine serum albumin. J. Colloid Interface Sci. 1981, 79, 548–565.
- Fullerton, G.D.; Kanal, K.M.; Cameron, I.L. Osmotically unresponsive water fraction on proteins: Non-ideal osmotic pressure of bovine serum albumin as a function of pH and salt concentration. Cell. Biol. Int. 2006, 30, 86–92.
- Rescic, J.; Vlacy, V.; Jamnic, A.; Glatter, O. Osmotic pressure, small-angle X-ray and dynamic light scattering studies of human serum albumin in aqueous solutions. J. Colloid Interface Sci. 2001, 239, 49–57.
- Jin, L.; Yu, Y.X.; Gao, G.H. A molecular-thermodynamic model for the interactions between globular proteins in aqueous solutions: Applications to bovine serum albumin (BSA), lysozyme, alpha-chymotrypsin, and immuno-gamma-globulin (IgG) solutions. J. Colloid Interface Sci. 2006, 304, 77–83.
- Giordano, R.; Grasso, A.; Wanderlingh, F.; Wanderlingh, U. Static and dynamic properties in thixotropic structures. Trends Colloid Interface Sci. V Prog. Colloid Polym. Sci. 1991, 84, 487–493.
- Soraruf, D.; Roosen Runge, F.; Grimaldo, M.; Zanini, F.; Schweins, R.; Seydel, T.; Zhang, F.; Roth, R.; Oettela, M.; Schreiber, F. Protein cluster formation in aqueous solution in the presence of multivalent metal ions – a light scattering study. Soft Matter 2014, 10, 894–902.
- Vekilov, P.G. Phase diagrams and kinetics of phase transitions in protein solutions. J. Phys. Condens. Matter 2012, 24, 193101.
- Dumetz, A.C.; Chockla, A.M.; Kaler, E.W.; Lenhoff, A.M. Protein phase behavior in aqueous solutions: Crystallization, liquid–liquid phase separation, gels, and aggregates. Biophys. J. 2008, 94, 570–583.
- Rozhkov, S.P.; Goryunov, A.S. Possible Phase Effects in the Dispersion of a Globular Protein in the Temperature Range of the Native State. Biophysics 2022, 67, 876–883.
- Sorkina, D.A. Heterogeneity of Serum albumin. Vopr. Med. Khimii 1991, 37, 14–17. (In Russian)
- Sugio, S.; Kashima, A.; Mochizuki, S.; Noda, M.; Kobayashi, K. Crystal structure of human serum albumin at 2.5 A resolution. Protein Eng. 1999, 12, 439–446.
- Carter, D.C.; He, X.-M. Structure of human serum albumin. Science 1990, 249, 302–303.
- Khachidze, D.G.; Monaselidze, D.R. Independent denaturation of albumin and globulin in human blood serum. Biophysics 2000, 45, 317–319.
- Stepuro, I.I.; Lapshina, E.A.; Chaikovskaja, N.A. Study of thermal denaturation of human serum albumin in water-alcohol and water-salt solutions in the presence of organic ligands. Molekularnajabiologia 1991, 25, 337–347. (In Russian)
- Turell, L.; Radic, R.; Alvarez, B. The thiol pool in human plasma: The central contribution of albumin to redox processes. Free Radic. Biol. Med. 2013, 65, 244–253.
- Stewart, A.J.; Blindauer, C.A.; Berezenko, S.; Sleep, D.; Tooth, D.; Sadler, P.J. Role of Tyr84 in controlling the reactivity of Cys34 of human albumin. FEBS J. 2005, 272, 353–362.
- Maier, R.; Fries, M.R.; Buchholz, C.; Zhang, F.; Schreiber, F. Human versus Bovine Serum Albumin: A Subtle Difference in Hydrophobicity Leads to Large Differences in Bulk and Interface Behavior. Cryst. Growth Des. 2021, 21, 5451–5459.
- Pavićević, A.; Luo, J.; Popović-Bijelić, A.; Mojović, M. Maleimidoproxyl as an EPR spin label for the evaluation of conformational changes of albumin. Eur. Biophys. J. 2017, 46, 773–787.
- Rezaei-Tavirani, T.; Moghaddamnia, S.H.; Ranjbar, B.; Amani, M.; Marashi, S.A. Conformational study of human serum albumin in predenaturation temperatures by differential scanning calorimetry, circular dichroism and UV spectroscopy. J. Biochem. Mol. Biol. 2006, 39, 530–536.
- Iosin, M.; Canpean, V.; Astilean, S. Spectroscopic studies on pH- and thermally induced conformational changes of Bovine Serum Albumin adsorbed onto gold nanoparticles. J. Photochem. Photobiol. A Chem. 2011, 217, 395–401.
- Borzova, V.A.; Markossian, K.A.; Chebotareva, N.A.; Kleymenov, S.Y.; Poliansky, N.B.; Muranov, K.O.; Stein-Margolina, V.A.; Shubin, V.V.; Markov, D.I.; Kurganov, B.I. Kinetics of thermal denaturation and aggregation of bovine serum albumin. PLoS ONE 2016, 11, e0153495.
- Gryzunov, Y.A.; Arroyo, A.; Vigne, J.L.; Zhao, Q.; Tyurin, V.A.; Hubel, C.A.; Gandley, R.E.; Vladimirov, Y.A.; Taylor, R.N.; Kagan, V.E. Binding of fatty acids facilitates oxidation of cysteine-34 and converts copper-albumin complexes from antioxidants to prooxidants. Arch. Biochem. Biophys. 2003, 413, 53–66.
This entry is offline, you can click here to edit this entry!
