1. Introduction
In the landscape of Parkinson’s Disease (PD) research, the fusion of wearable sensor data with personal health records (PHR) has emerged as a pivotal avenue, promising to enhance patient monitoring and alerting capabilities.
This study delves into the intricate domain of PD patient care, with a specific emphasis on harnessing the potential of wearable sensors to capture, represent, and semantically analyze PD patient’s movement data and domain knowledge. The primary objective is to elevate the assessment of PD patients by establishing a robust foundation for personalized health insights through the development of Personal Health Knowledge Graphs (PHKGs)
[1][2][3]. Additionally, a personal health Graph Neural Network (PHGNN) is developed leveraging the PHKG to formalize the representation of related sensors and PHR integrated/unified data at a higher level of abstraction
[4]. This paper, as an extension of our previous related work
[5], provides a detailed exploration of the Wear4PDmove ontology and evaluates its integration within the development of an experimental PHKG
[5]. Furthermore, the focus is on integrating PHKG within PHGNN implementations, emphasizing the importance of comprehensive PD-related data integration for effective patient monitoring and alerting. To achieve this goal, we have implemented a rule-based notification mechanism on specific features from PHKG with two different levels of notification (medium and high). These notifications provide health experts with precise and timely information and contribute to the continuous evaluation of PD patient-related events. This ultimately enhances patient care by facilitating well-informed time and life-critical medical decision-making.
The challenge lies in seamlessly integrating off-the-shelf wearable sensor data with PHRs to create a comprehensive and interoperable framework for effective PD patient monitoring and alerting
[6]. This requires addressing complexities in data representation, interoperability, and high-level event recognition. The main target of the approach is to propose a holistic solution for personalized PD patient monitoring and alerting in terms of the Wear4PDmove ontology applied over a novel experimental PHKG framework, which is appropriately integrated with PHGNNs.
The motivation stems from the need to enhance the monitoring, assessment and alerting of PD patients by creating an advanced knowledge representation and reasoning (KRR) system. Integrating off-the-shelf wearable sensor data and PHRs into a PHKG along with PHGNNs
[7] aligns with the growing demand for sophisticated and personalized healthcare solutions.
The key contribution of the work presented in this paper, extending our previous related work, is:
-
Experimental PHKG: Evaluation of the Wear4PDmove ontology within the development of an experimental PHKG, showcasing its practical applicability.
-
PHKG integration with GNNs: Emphasis on the integration of PHKG within GNN implementations, elucidating its pivotal role in advancing PD monitoring and alerting solutions.
-
Precision in PD Monitoring: Addressing specific demands in PD monitoring through the precision offered by the Wear4PDmove ontology.
-
Enhanced Data Analysis with GNNs: Implementation of GNNs to facilitate enhanced analysis of complex medical data.
-
Capturing Intricate Relationships: Exploration of GNNs’ capabilities to capture intricate relationships within PD-related data, enhancing understanding and insights.
-
Comprehensive PHKG: Creation of a comprehensive PHKG integrating diverse data sources, including movement data, clinical records, and health-related information.
The overall contributions of our work in this line of research are as follows:
-
Interoperability and Interlinkage: Ensuring PHKG interoperability and seamless interlinkage with relevant datasets, promoting comprehensive data utilization.
-
Real-Time Alerting Mechanisms: Implementation of real-time alerting mechanisms utilizing rule-based high-level event recognition, contributing to timely interventions in PD care.
-
Advanced Semantic Data Analysis: Development of advanced semantic data analysis techniques for uncovering movement data semantics, contributing to nuanced insights.
-
Patient-Centric Solutions: Contribution to practical, patient-centric solutions for improved PD monitoring and care, emphasizing the integration of technology for personalized healthcare.
The structure of this paper is as follows:
Section 2 provides essential background knowledge on the subject. In
Section 3, we present a curated selection of state-of-the-art scientific studies.
Section 4 delves into the detailed presentation of the Wear4PDmove ontology, elucidating its significance in capturing and formalizing PD-related data.
Section 5 explores the evaluation of the ontology within the development of an experimental PHKG, showcasing its practical applicability and interoperability, along with an examination of its integration within GNNs. Moving beyond the state-of-the-art,
Section 6 initiates a comprehensive discussion on the limitations and open issues associated with the Wear4PDmove ontology, providing insights into potential challenges and avenues for future research. Finally,
Section 7 concludes the paper and outlines plans for future research.
2. Background Knowledge
This section presents key concepts spanning symbolic artificial intelligence (AI) and ontological frameworks, Knowledge Graphs (KG), PHKG, and Neurosymbolic AI
[8]. Symbolic AI, epitomized by ontologies, forms the cornerstone of logical reasoning and knowledge representation, laying the groundwork for understanding/capturing complex relationships within healthcare data. KGs, through their capacity to organize and contextualize information, offer reasoning capabilities that result in valuable insights. We extend this paradigm to the case of PHKG, interlinking and reasoning with diverse health-related datasets. The emergence of Neurosymbolic AI, coupling KGs with neural networks into GNNs, signifies a novel approach to decoding intricate medical data. This collective exposition establishes the essential backdrop for our subsequent in-depth exploration and contributions in the following sections.
2.1. Symbolic AI and Ontology Engineering
Symbolic AI, encompassing logical reasoning, knowledge representation, and rule-based systems, constitutes a multifaceted approach to artificial intelligence. At its core, logical reasoning forms the basis of AI systems, employing several types of mathematical logic to support deductive reasoning and decision-making mechanisms. Knowledge representation involves the explicit portrayal of information using symbols, rules, and relationships
[9]. This structured representation allows systems to comprehend intricate datasets effectively and manipulate them.
In Internet of Things (IoT) environments, ensuring interoperability is challenging due to the heterogeneous nature of data from devices like health and fitness trackers. Proprietary formats and lack of common terms often lead to certain challenges related to communication protocols and data formats
[10]. Overcoming these challenges requires adopting common syntax and semantics for IoT-generated data. The Semantic Web of Things (SWoT) technology has emerged as a solution to mitigate such data and information heterogeneity. Semantics, dealing with meaning and truth, are harnessed through the Resource Description Framework (RDF) for semantically annotated linked data
[11]. Semantic models and standards, such as the Semantic Sensor Network (SSN)
[12], Sensor, Observation, Sample, and Actuator (SOSA)
[13], Smart Applications Reference Ontology (SAREF)
[14], and Data Analytics for Health and Connected Care (DAHCC) ontology
[15], address the challenge of bridging sensor data heterogeneity. The Parkinson Movement Disease Ontology (PMDO)
[16] specifically caters to Parkinson’s Disease, organizing information into neurological findings, treatment plans, and evaluation instruments. Utilizing SW technologies, particularly RDF data modeling, facilitates semantic interoperability in IoT-enabled health devices
[17].
Ontologies play a key role in semantic modeling, explaining object behavior and enriching raw sensor data. While progress has been made in developing semantic models for healthcare, there is an ongoing interest in developing models focusing on sensor data related to PD and wearable devices
[18]. Health-related ontologies serve as conceptual models providing structure and meaning to health data, fostering common formats for knowledge exchange among humans and software agents involved in healthcare applications and systems.
Considering symbolic AI, the development of rule-based systems is commonplace, where sets of logical rules govern the system’s behavior
[19]. These rules, derived from data-driven learning strategies, serve as the guiding principles for drawing conclusions from given inputs. Technologies like RDF and OWL
[20], often employed alongside Symbolic AI methodologies, play a crucial role in effective implementation.
2.2. Knowledge Graphs
KG serves as a structured and interconnected representation of information, adopting a graph format to represent real-world entities, their attributes, and their relationships
[21]. This semantic framework enhances understanding and facilitates reasoning. KGs feature a graph structure comprising nodes (representing entities) and edges (representing relationships), allowing the depiction of complex inter-entity connections. By incorporating Linked Data (LD) principles, KGs interconnect with external sources of information, thereby enhancing the comprehensiveness of information
[22]. Their scalability accommodates vast datasets with efficient traversal and retrieval capabilities.
KG applications include semantic search (e.g., Google search), recommendation systems, data integration, and support for knowledge-based AI. RDF is fundamental to KGs, using triples (subject-predicate-object) to express statements. SPARQL (SPARQL Protocol and RDF Query Language)
[23] facilitates the querying and manipulation of RDF data within a KG.
2.3. PHKG
In the evolving landscape of healthcare informatics, the development and utilization of PHKGs have emerged as instrumental components for advancing personalized medicine. A PHKG represents a structured and interconnected knowledge base that combines diverse personal health data, ranging from clinical records to sensor-derived information, with the aim of fostering a holistic understanding of an individual’s health profile
[24]. The conceptual underpinning of a PHKG draws from the principles of the Semantic Web (SW) and LD standards, facilitating the creation of a comprehensive framework that transcends traditional health record silos.
Central to the efficacy of a PHKG is the integration of various application and domain-specific ontologies, which serve as formalized structures defining relationships and hierarchies of entities within the graph. Specifically, in the context of PD, a well-defined ontology, such as the Wear4PDmove ontology, plays a pivotal role in ensuring precision and specificity in representing and reasoning with PD-related data. The semantic richness afforded by these ontologies not only aids in the interoperability of diverse health data sources but also forms the basis for rule-based event recognition, enabling timely and contextually relevant medical interventions
[25]. As wearable sensor technology becomes increasingly prevalent in health monitoring, the synergy of ontologically structured data within a PHKG stands as a promising paradigm for tailoring healthcare insights to the unique nuances of an individual’s health journey
[2].
2.4. Neurosymbolic AI and GNNs
Neurosymbolic AI is a powerful interdisciplinary approach at the intersection of symbolic reasoning and neural network-based learning, aiming to harness the logical strengths and explicit knowledge representation of symbolic methods and the pattern recognition capabilities of neural networks (NNs). This hybrid model seamlessly integrates symbolic reasoning, often associated with rule-based systems, with subsymbolic learning, utilizing neural networks for effective pattern recognition
[26]. One of its distinguishing features is the capability to create interpretable and explainable AI models by leveraging explicit rule-based representations from symbolic components. A key subset, GNNs, excels in processing graph-structured data by capturing intricate dependencies and interactions among entities (nodes) connected by relationships (edges). GNNs use a message-passing mechanism, allowing nodes to iteratively exchange information and aggregate insights, deepening their understanding of the graph’s structure.
In diverse applications, neurosymbolic AI and GNNs play a crucial role. In drug discovery, they combine expert knowledge with GNN-driven insights to accelerate the identification of potential drug candidates
[27]. In healthcare, these models enhance the interpretability of medical decision-making by providing explanations for diagnoses through the integration of symbolic medical knowledge with learned patterns from patient data
[28]. Social network analysis benefits from GNNs’ ability to capture relationships and influence patterns within graphs, further enriched by incorporating symbolic reasoning for a comprehensive understanding of social structures.
In the context of PD research, data analysis assumes a pivotal role, leveraging Machine Learning (ML) and Deep Learning (DL) techniques for comprehensive exploration. Datasets, including electronic health records (EHRs), patient-reported outcomes, and imaging data, play a crucial role in understanding PD’s mechanisms. The integration of KGs and GNNs emerges as a significant advancement, organizing PD-related information and excelling in tasks like patient classification and disease progression prediction. The application of ML, GNNs, and DL contributes to uncovering hidden patterns, enhancing treatment modalities, and refining therapeutic strategies in PD research.
3. Related Work
In the realm of PD research, a comprehensive understanding relies on the exploration of various domains, including ontologies, PHKGs, and GNNs. This section delves into state-of-the-art research, examining the pivotal role of ontologies in structuring PD-related knowledge, the application of PHKGs for personalized health insights, and the advancements brought by GNNs in analyzing complex medical data. Each sub-section within this segment unfolds the landscape of relevant studies, providing insights into the contributions, methodologies, and findings that have shaped the understanding of PD through ontological frameworks, KGs, and NN approaches.
3.1. Ontologies for PD
An ontology in the health domain serves as a framework for organizing and adding meaning to health-related data and information that is shared across different applications, services, and systems. Such ontologies play a crucial role in representing, integrating, and sharing health-related knowledge in a standardized and widely accepted format that can be understood by both humans, such as doctors and patients, and software agents. Younesi et al.
[1] have developed the PD Ontology (PDON) as a standardized vocabulary and definition system for PD and its associated symptoms, treatments, and clinical studies. The ontology has been built using standard ontology development life cycle stages such as requirements gathering, conceptualization, implementation, and evaluation. PDON is designed to support interoperability and data sharing in the PD research community and includes a subclass-based taxonomic hierarchy that spans from molecular to clinical concepts. The evaluations have demonstrated that PDON can answer specific PD-related questions and improve the accuracy and consistency of data annotation and analysis.
The Parkinson’s Movement Disorder Ontology (PMDO)
[29] is a significant ontology for PD created by movement disorder specialists. The PMDO encompasses three major categories: neurological observations, treatment plans, and tools used to evaluate various aspects of PD. This ontology is a valuable asset for researchers and clinicians to label, share, and consolidate data related to PD and other movement disorders, as it offers a uniform terminology for these conditions. Overall, the PMDO is an important resource for the PD research community that can facilitate data sharing and interoperability and aid in standardizing the analysis and interpretation of PD-related information. PMDO is a more up-to-date resource than PDON, as the latter was last updated in 2016. Both ontologies are accessible through NCBO BioPortal
[30], providing researchers and clinicians with standardized vocabularies for annotating and sharing PD-related data. While both the PMDO and PDON ontologies provide a standardized vocabulary and definitions for various aspects of PD and related movement disorders, neither ontology contains specific conceptualizations for representing knowledge extracted from data collected via wearable sensors. This is a limitation of both ontologies, as the use of wearable sensors is becoming increasingly common in PD monitoring and clinical care. Wearable sensors provide a way to collect objective and quantitative data on PD symptoms and motor function on a daily basis outside of a clinical setting.
3.2. PHKGs for PD
Recent studies have investigated the advantages of PHKGs in advancing smart health applications. A representative example is the IoT Semantic Annotations System (IoTSAS)
[31], which processes real-time sensor stream data, integrating semantic annotations to deliver immediate health information to citizens, especially concerning air pollution and weather conditions. Another study
[32] utilized data from multiple IoT fitness vendors to establish a context-aware resource graph linked to health-related ontologies. Through a web portal, they facilitated the integration, sharing, and analysis of diverse IoT health and fitness data. Additionally,
[33] proposed an OWL-based integration, combining ontologies to model health data and Web of Things (WoT) services, successfully creating a machine-understandable and cross-system reusable resource graph by linking health and home environment data. Notably, despite semantic annotation exploration in related work, there is a notable gap as no studies have developed PHKGs integrating real-time sensor data from wearables and other PHR sources for monitoring and alerting in PD patients.
Ontologies offer a means to explicitly represent unified knowledge related to sensor and PHR data, supporting knowledge creation through semantic inferencing. While advancements have occurred in semantic models for healthcare monitoring, there remains untapped potential for developing or extending existing models, particularly those focused on rules-based high-level event recognition for PD monitoring and alerting.
3.3. GNNs for PD
GNNs rapidly advance PD research through robust data analysis, handling patient-reported outcomes, imaging data, and EHRs. Integrated into knowledge graphs, GNNs unveil intricate patterns, offering insights into PD’s mechanisms. Progress in GNNs for PD diagnosis, particularly Deep Learning models analyzing imaging data, shows potential for detection and prognosis. These advancements aim to enhance patient well-being and alleviate PD-related healthcare burdens, holding promise for future exploration and improved understanding.
Recent advances in GCNs, particularly in PD diagnosis using Deep Learning models for imaging data scrutiny, are promising in enhancing patient outcomes and reducing the healthcare burden
[34]. The Multi-View Graph Convolutional Network (MV-GCN) enhances prediction accuracy in PD-related predictive tasks, utilizing multiple brain graph inputs
[35][36]. Validation with real-world data from the Parkinson’s Progression Markers Initiative (PPMI) demonstrates its promising performance in predicting pairwise matching relationships in the context of PD
[37].
ML and DL techniques for identifying blood-based biomarkers in Alzheimer’s and PDs, especially leveraging Convolutional Neural Networks (CNNs), show potential for early diagnosis and clinical trial screening
[38][39]. The field of neuroimage analysis witnesses the popularity of GCNs in diverse applications, including distinguishing cases and controls in autism research.
Notably, GATs excel in managing extensive and intricate graphs, effectively filtering irrelevant data during DP. Advances in GAT applications for PD diagnosis, employing DL models for imaging data analysis, show promise in enhancing accuracy rates and patient outcomes
[40]. Studies introduce novel approaches like multimodal GCN (M-GCN) and GAT models for predicting phenotypic measures, showcasing ongoing exploration of graph-based NN models to advance understanding neurological conditions
[41]. This study proposes a deep multi-modal fusion model (DMFM) based on GAT, effectively incorporating spatial dependencies for spatiotemporal correlation modeling using ConvLSTM and a temporal attention mechanism (TAM)
[42], ultimately enhancing prediction accuracy.
Recent advances in deploying GRNs for PD diagnosis, particularly using DL models for analyzing time-series imaging data, show promising results with high accuracy rates
[43]. This suggests the potential to enhance patient outcomes and alleviate the burden of PD on healthcare systems. In summary, GRNs offer a robust tool for processing time-series data in PD research, holding substantial promise for advancing our understanding of this complex condition and improving patient outcomes
[44].
GTNs demonstrate proficiency in managing complex graph structures, particularly relevant in handling intricate data sources like EHRs and imaging data. This enables GTNs to capture the intricate interconnections between various data elements, providing a comprehensive perspective on disease progression and patient outcomes. Recent advancements in applying GTNs to PD diagnosis and treatment involve the use of DL models for analyzing imaging data and predicting disease progression
[45].
GAEs also serve as valuable tools for dimensional reduction, addressing challenges posed by high-dimensional datasets in PD research, especially in imaging data, and facilitating the identification of latent patterns and relationships. Significant progress has been achieved in employing GAEs for PD diagnosis and therapeutic interventions, utilizing DL models for analyzing imaging data and predicting disease progression
[46].
In conclusion, GAEs emerge as vital and adaptable tools in PD research due to their competence in managing graph-structured data and simplifying complex datasets through dimensionality reduction, positioning them as valuable assets in unraveling the intricacies of PD. Furthermore, in the broader scientific landscape, innovative methods like DD-GCN and SLMGAE are introduced for predicting human splice-site events and protein–protein interactions, respectively
[47]. DD-GCN employs a two-pronged dropout strategy for enhancing gene embeddings, while SLMGAE utilizes an autoencoder architecture for obtaining low-dimensional graph representations
[48].
By combining graph-based representations and generative models, GGNs offer a powerful tool for unraveling the intricate relationship between PD symptoms and disease pathology, paving the way for more effective and personalized PD treatments. In the realm of medical image analysis
[49], GGNs play a crucial role in data augmentation, addressing limitations in labeled images by employing a generative framework with a generator network crafting synthetic data and a discriminator network distinguishing between real and synthetic data.
The integration of graph-based representations with reinforcement learning enhances the precision of treatment choices for PD patients, resulting in improved outcomes. Notable applications of deep reinforcement learning networks in the medical field include predicting brain tumor locations using a deep Q-network (DQN)
[49], a method for medical image semantic segmentation, and a recommendation system for antihypertensive medications for patients with hypertension and type 2 diabetes
[50].
4. The Wear4PDmove Ontology
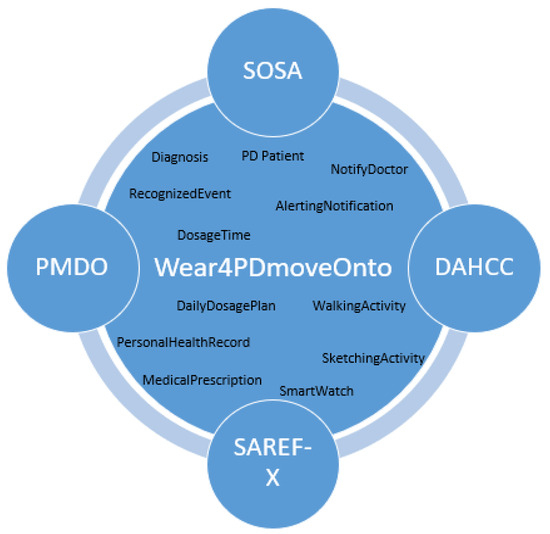
The ontology proposed in this work represents knowledge related to the monitoring of PD patients’ movement and to the integration of such data with PHR data towards supporting real-time recognition of events such as a missing dose event, eventually triggering the appropriate alerting. Wear4PDmove is an ontology that reuses and extends other ontologies, including DAHCC, SOSA, SAREF, and PMDO (Figure 1). The imported ontologies extend the capabilities of Wear4PDmove ontology by providing the necessary concepts and relationships to model real-world data and knowledge related to PD patients’ monitoring and alerting.
This entry is adapted from the peer-reviewed paper 10.3390/info15020100