Mitochondrial ATP synthase (Complex V) catalyzes the last step of oxidative phosphorylation and provides most of the energy (ATP) required by human cells. The mitochondrial genes MT-ATP6 and MT-ATP8 encode two subunits of the multi-subunit Complex V. Since the discovery of the first MT-ATP6 variant in the year 1990 as the cause of Neuropathy, Ataxia, and Retinitis Pigmentosa (NARP) syndrome, a large and continuously increasing number of inborn variants in the MT-ATP6 and MT-ATP8 genes have been identified as pathogenic. Variants in these genes correlate with various clinical phenotypes, which include several neurodegenerative and multisystemic disorders.
- mitochondria
- ATP synthase
- ATP6
- ATP8
- mt-DNA
1. Introduction
| Genetic Variant/Subunit AA Change | Tissue or Cell Models H (%) |
Biochemical and Cellular Parameters | ||||
|---|---|---|---|---|---|---|
| CV ATP Synthesis |
CV ATP Hydrolysis |
OCR | CV Assembly/Stability | Other Mitochondrial and Cellular Readouts |
||
| m.8382C>T ATP8: p.T6I |
Muscle (100%) [3] |
(D) | CI activity (D) | |||
| Fibroblasts (100%) [3] |
(N) | (N) | CIV activity (D) | |||
| m.8403T>C ATP8: p.I13T |
Fibroblasts (100%) |
(N) [3] | (N) [3] | Depolarized plasma membrane and ROS (I) [4]; CIV activity (D) [3] |
||
| Yeast (100%) [5] |
(D) | (N) | Growth in stress conditions (D); Mitochondrial membrane potential (N) |
|||
| m.8424T>C ATP8: p.I20P |
Muscle (100%) [3] |
(D) | CI, CII, CIII, and CIV activities (D) | |||
| Fibroblasts (100%) [3] |
(D) | (N) | CI activity and growth in galactose media (D) | |||
| Cybrids (100%) [3] |
(D) | (D) | CI and CIV activities (D); Lactate production (I) |
|||
| m.8528T>C ATP8: p.W55R ATP6: p.M1T |
Fibroblast (93%) [6] |
(D) | ||||
| Heart muscle (90%) [7] | (A) | CV subunit levels and CI activity (D); ATP6 and ATP8 protein levels (D) |
||||
| m.8529G>A ATP8: p.W55X ATP6: p.M1M |
Muscle (>90%) [8] |
(D) | (D) | (A) | CI-CIV activities (N) | |
| Fibroblast (>90%) [8] |
(D) | CI-CIV activities (N) | ||||
| Cybrids (100%) |
(D) [8][9] | (D) [9] | (A) [8][9] | Growth in galactose media (D); ATP6 and ATP8 protein levels (D); Complexes II, III, and IV levels (D) [9] |
||
| m.8561C>G ATP8: p.P66A ATP6: p.P12R |
Myoblasts (99%) [10] |
(A) | Total ATP level (D); ROS (N); ATP6 and ATP8 protein levels (N) |
|||
| m.8561C>T ATP8: p.P66L ATP6: p.P12S |
Muscle (99%) [11] | (D) | (A) | |||
| m.8611insC ATP6: p.L29PfsX36 |
Muscle (60%) [12] |
(D) | (A) | |||
| Fibroblasts (80%) [12] |
(D) | (A) | ATP6 protein level (D); Mitochondrial cristae structure and dynamics (A) |
|||
| m.8618insT ATP6: p.T33HfsX32 |
Muscle (65–85%) |
(A) [13][14] | ATP6 protein level (D) [13] | |||
| Fibroblasts (45%) [14] |
(D) | (A) | ROS (I); Mitochondrial network morphology (N) |
|||
| m.8648G>A ATP6: p.R41Q |
Fibroblast (100%) [3] |
(N) | (N) | |||
| Cybrids (100%) [3] |
(N) | (N) | ||||
| m.8782G>A ATP6: p.G86X |
Fibroblasts (12–27%) [14] |
(D) | (A) | ROS (I); Mitochondrial morphology (N) |
||
| m.8806C>G ATP6: p.P94A |
Muscle (100%) [3] |
(N) | CI-CIV activities (D) | |||
| m.8839G>C ATP6: p.A105P |
Cybrids (100%) [15] |
(N) | Growth in galactose media (D); Mt-DNA copy number (I); OXPHOS protein levels (I); Mitochondrial membrane potential (D); CI-CIV activities (N) |
|||
| m.8843T>C ATP6: p.I106T |
Yeast (100%) [16] |
(N) | (N) | (N) | Mitochondrial membrane potential (N) | |
| m.8851T>C ATP6: p.W109R |
Yeast (100%) [17] |
(D) | (D) | (D) | (N) | Growth in stress conditions (D); Mitochondrial cristae structure (A); CIII and CIV super-complexes (D) |
| m.8909T>C ATP6: p.F128S |
Yeast (100%) [18] |
(D) | (D) | (A) | ||
| m.8932C>T ATP6: p.P136S |
Yeast (100%) [19] |
(D) | (D) | (A) | ATP6 protein level (D) | |
| m.8946A>C ATP6: p.M140I |
Fibroblasts (100%) [3] |
(N) | (N) | CI activity (D) | ||
| m.8950G>A ATP6: p.V142I |
Lymphocytes [20] | (D) | ||||
| Yeast (100%) [16] | (D) | (D) | (N) | Sensitivity of growth to oligomycin (I); Mitochondrial membrane potential (N) |
||
| m.8969G>A ATP6: p.S148N |
Muscle (100%) [3] | (N) | CI activity (D) | |||
| Yeast (100%) |
(D) [21][22] | (D) [21] | (D) [21][22] | (A) [21] | Growth in stress conditions (D) [21][22] | |
| Cybrids (19–98%) [21] |
(D) | Mitochondrial cristae structure (A); ROS (I) |
||||
| Fibroblasts (100%) [23] |
(D) | |||||
| m.8975T>C ATP6: p.L150P |
Muscle [3] | (D) | CI activity (D) | |||
| Fibroblasts (100%) [3] |
(D) | (N) | CI activity (D); Growth in galactose media (D) |
|||
| Cybrids (100%) [3] |
(N) | (N) | CI and CIV activities (D); Growth in galactose media (D); Lactate production (I) |
|||
| m.8989G>C ATP6: p.A155P |
Muscle (92%) [24] |
(D) | Mitochondrial ultrastructure (N) | |||
| m.8993T>G ATP6: p.L156R |
Yeast (100%) [25] |
(D) | (D) | (D) | (A) | Growth in stress conditions (D); CIV level (D) |
| Platelets (80–93%) |
(D) [26][27] | (N) [26][27] | CV ATP-driven proton flow (N) [26] | |||
| Lymphocytes (80–100%) |
(D) [28][29][30][31] | (D) [28] | (D) [29] | ROS and mitochondrial membrane potential (I) [31]; CV proton flow (D) [30][32]; Oligomycin sensitivity of CV proton flow (I) [32] |
||
| Muscle (76%) [33] |
(A) | |||||
| Fibroblasts (70–100%) |
(D) [3][34][35][36][37][38][39] | (D) [35][38][40]; (N) [3][34][36] | (D) [41][42]; (N) [34] |
(N) [36][37] | Mitochondrial membrane potential (I) [36][39]; Mitochondrial morphology (A) [39][42]; ROS (I) [39][43]; Antioxidant enzymes (A) [39]; Oligomycin sensitivity of CV (I) [34]; Growth in galactose media (D) [3][35][38][40]; Mitochondrial calcium uptake (D) [39]; Glycolytic capacity (D) [42]; CI and CIV activities (D) [3] |
|
| Cybrids (45–100%) |
(D) [3][35][37][40][44][45][46][47][48][49] | (D) [3]; (N) [40] |
(D) [29][40][46][47][48][50][51] | (A) [37][44][48] | Mitochondrial membrane potential (D) [50] or (I) [47][49]; Mitochondrial morphology (A) [52][53]; Mitochondrial ultrastructure (A) [49]; ROS (I) [47][50][54]; Antioxidant enzymes (A) [47][54]; Growth in galactose media (D) [35][45][47]; ATP level (D) [50]; Extracellular lactate (I) [3][46]; Autophagy (I) [53]; CI, CII, or CIV activities (D) [3][47][48]; Oligomycin [37] and apoptosis [49] sensitivity (I); Actin cytoskeleton and Ca2+ in-flux rates (A) [52]; Reductive carboxylation of glutamine and NADH/NAD ratio (I) [51][55] |
|
| IPSCs (90–100%) |
(D) [56]; (N) [57] | Mitochondrial membrane potential, ROS, and lactate production (I) [58] | ||||
| NPCs, Neurons (90–100%) |
(D) [58] | Mitochondrial membrane potential, ROS, and antioxidant enzymes (I) [58]; Degenerative defect [58]; Metabolic dysregulation; Formation of cerebral organoid (A) [57] |
||||
| m.8993T>C ATP6: p.L156P |
Yeast (100%) [59] |
(D) | (N) | (D) | (N) | CIV level, COX2, and ATP6 protein levels (D) |
| Lymphocytes (90–95%) |
(D) [31] | Mitochondrial membrane potential (N); ROS (I) [31]; Proton flux (D) [32] |
||||
| Fibroblasts (95–100%) |
(D) [3][34] | (D) [3], (N) [34] | (N) [34] | (N) [37] | Depolarized plasma membrane and ROS (I) [4]; Growth in galactose media (D) [3] |
|
| Cybrids (100%) |
(D) [40][46]; (N) [3][48] |
(N) [3] | (D) [46]; (N) [48] |
(N) [37][48] | Lactate production (I) [3][46] | |
| m.9008C>G ATP6: p.T161S |
Muscle (100%) [3] |
(N) | ||||
| Fibroblasts (100%) [3] |
(N) | (N) | CI activity (D) | |||
| Cybrids (100%) [3] |
(D) | (N) | Growth in galactose media (D); Lactate production (I) |
|||
| m.9016A>G ATP6: p.I164V |
Yeast (100%) [16] |
(N) | (N) | (N) | Mitochondrial membrane potential (N) | |
| m.9019A>G ATP6: p.T165A |
Muscle (100%) [3] |
(D) | CI activity (D) | |||
| m.9025G>A ATP6: p.G167S |
Yeast (100%) [16] |
(D) | (D) | (N) | Sensitivity of growth to oligomycin (I); Mitochondrial membrane potential (N) |
|
| m.9029A>G ATP6: p.H168R |
Yeast (100%) [16] |
(D) | (D) | (N) | Sensitivity of growth to oligomycin (I); Mitochondrial membrane potential (N) |
|
| Cybrids (100%) [50] |
(D) | ATP level (D); ROS and mitochondrial membrane potential (I) |
||||
| m.9032T>C ATP6: p.L169P |
Cybrids (25–80%) [50] |
(D) | ATP level (D); ROS and mitochondrial membrane potential (I) |
|||
| m.9035T>C ATP6: p.L170P |
Cybrids (100%) [60] |
(D) | ROS and antioxidant enzymes (I); Mitochondrial membrane potential (N); Sensitivity to glucose deprivation (I); Oxidative stress (I) |
|||
| Muscle (100%) [3] |
(D) | CI activity (D) | ||||
| Fibroblasts (100%) |
(D) [3] | (N) [3] | (D) [61] | (A) [61] | Growth in galactose media (D) [3] | |
| m.9058A>G ATP6: p.T178A |
Yeast (100%) [16] |
(N) | (N) | (N) | Mitochondrial membrane potential (N) | |
| m.9101T>C ATP6: p.I192T |
Lymphocytes (100%) [62][63] |
(D) | ||||
| Cybrids (100%) [63] |
(D) | |||||
| m.9127 delAT ATP6: p.I201PfsX2 |
Fibroblasts (50%) [64] |
(D) | (D) | (N) | Oligomycin-induced increase in mitochondrial membrane potential (D) | |
| m.9134A>G ATP6: p.E203G |
Muscle [65] | (D) | (D) | |||
| m.9139G>A ATP6: p.A205T |
Yeast (100%) [16] |
(N) | (N) | (N) | Mitochondrial membrane potential (N) | |
| m.9154C>T ATP6: p.Q210X |
Fibroblasts [66] | (N) | (A) | Mitochondrial morphology (A) | ||
| IPSC and Neurons [66] | (A) | Motor neuron differentiation (A); Mitochondrial morphology (A); Hyperactivation of the Notch pathway |
||||
| m.9160T>C ATP6: p.Y212H |
Yeast (100%) [16] |
(N) | (N) | (N) | Mitochondrial membrane potential (N) | |
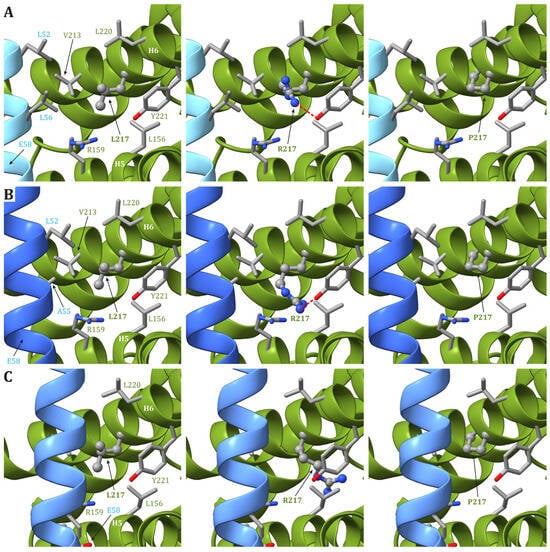
| m.9176T>G ATP6: p.L217R |
Yeast (100%) [67] |
(D) | (D) | (A) | Growth in stress conditions (D); CIV super-complexes (D); ATP6, COX2, and CYTB protein levels (D); Mitochondrial ultrastrucure (A) |
|
| Muscle (>95%) [33] |
(A) | |||||
| Fibroblasts (95–100%) |
(D) [68] | (N) [40] | (N) [68] | Mitochondrial membrane potential (I) [68]; Growth in galactose media (D) [40] |
||
| Cybrids (30–100%) |
(D) [40][48][49] | (N) [40] | (D) [40][48] | (A) [48] | CI and CIV activities (D) [48]; Mitochondrial ultrastructure (A) [49]; Mitochondrial membrane potential and apoptosis sensitivity (I) [49] |
|
| m.9176T>C ATP6: p.L217P |
Yeast [69] | (D) | (N) | (D) | (A) | |
| Muscle (100%) [3] |
(D) | |||||
| Fibroblasts (100%) |
(N) [70]; (D) [71] | (N) [40] | (A) [71] | Mitochondrial network morphology (N) [71]; Depolarized plasma membrane and ROS (I) [4] |
||
| Cybrids (100%) [40] |
(D) | (N) | ||||
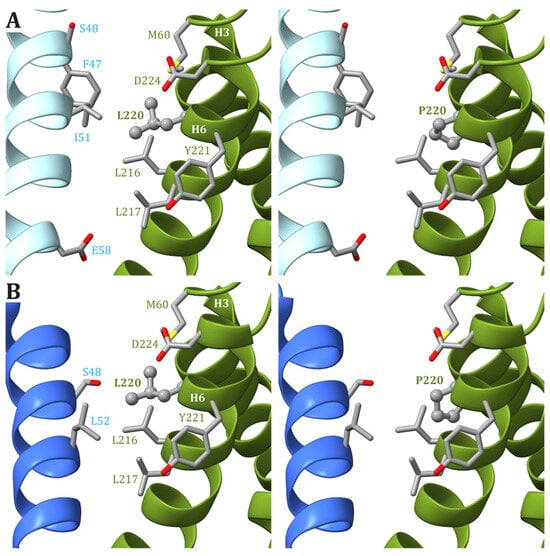
| m.9185T>C ATP6: p.L220P |
Yeast [72] | (D) | (N) | (N) | (N) | Sensitivity of growth to oligomycin (I) |
| Muscle (>97%) |
(D) [73][74] | (A) [74] | CI, CII, and CIV activities (N) [75][76][77] | |||
| Lymphocytes [75] |
(N) | (D) | ||||
| Fibroblasts (90–100%) |
(D) [3][4] | (D) [4]; (N) [3] | (D) [4] | (N) [4] | CI activity (D) and depolarized plasma membrane [4]; ROS or antioxidant enzymes (I) [4][78]; CI, CII, and CIV activities (N) [75]; Mitochondrial membrane potential (N) [78]; Lactate production (I) [3] |
|
| Cybrids (100%) | (D) [78]; (N) [3] |
(D) [4]; (N) [3] | (D) [4] | CI activity (D) [4]; Lactate production (I) [3]; Mitochondrial membrane potential (N) [78] |
||
| NPC and neuron (100%) [78] | (D) | (N) | Mitochondrial membrane potential (I); Mitochondrial calcium homeostasis (A); Depolarized plasma membrane; Mitochondrial cristae structure and ROS (N) |
|||
| m.9191T>C ATP6: p.L222P |
Muscle (94%) [73] |
(D) | (D) | |||
| Yeast [72][79] |
(D) | (N) | (D) | (A) | Growth in stress conditions (D); CIV level (D); ATP6 protein level (D) |
|
| m.9205delTA ATP6: p.X227NA |
Muscle (>98%) [80] |
CIV activity (D) | ||||
| Fibroblasts (>98%) |
(D) [80] | (N) [80] | (D) [80] | (A) [80] | CIV activity (D) [80][81]; ATP6 protein and CIV subunit levels (D) [80]; Morphological abnormalities of mitochondria [81] |
|
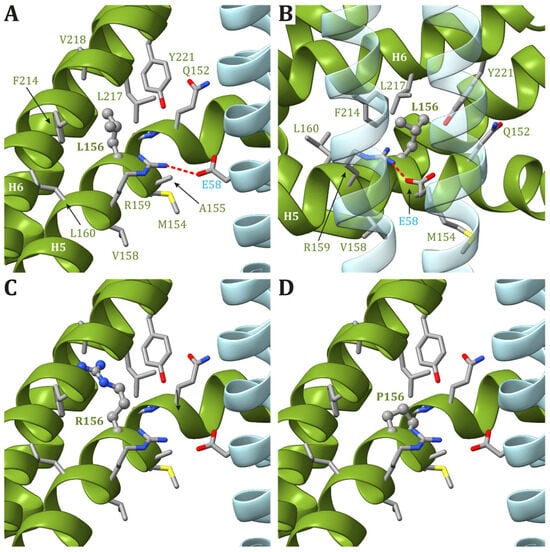
2. The mt-DNA Pathogenic Variants at Position m.8993
2.1. Biochemical and Cellular Dysfunctions in Mutated Cell Models
2.2. Modeling of ATP6 Subunit Carrying Changes in Leu156 in the ATP Synthase Human Structure
3. The Pathogenic Variants at Nucleotide m.9176
4. The mt-DNA Pathogenic Variant at Position m.9185
5. Other MT-ATP6 and MT-ATP8 Pathogenic Variants
This entry is adapted from the peer-reviewed paper 10.3390/ijms25042239
References
- Gustafsson, C.M.; Falkenberg, M.; Larsson, N.-G. Maintenance and Expression of Mammalian Mitochondrial DNA. Annu. Rev. Biochem. 2016, 85, 133–160.
- Picca, A.; Guerra, F.; Calvani, R.; Coelho-Júnior, H.J.; Leeuwenburgh, C.; Bucci, C.; Marzetti, E. The Contribution of Mitochondrial DNA Alterations to Aging, Cancer, and Neurodegeneration. Exp. Gerontol. 2023, 178, 112203.
- Rucheton, B.; Jardel, C.; Filaut, S.; Amador, M.D.M.; Maisonobe, T.; Serre, I.; Romero, N.B.; Leonard-Louis, S.; Haraux, F.; Lombès, A. Homoplasmic Deleterious MT-ATP6/8 Mutations in Adult Patients. Mitochondrion 2020, 55, 64–77.
- Aure, K.; Dubourg, O.; Jardel, C.; Clarysse, L.; Sternberg, D.; Fournier, E.; Laforet, P.; Streichenberger, N.; Petiot, P.; Gervais-Bernard, H.; et al. Episodic Weakness Due to Mitochondrial DNA MT-ATP6/8 Mutations. Neurology 2013, 81, 1810–1818.
- Panja, C.; Niedzwiecka, K.; Baranowska, E.; Poznanski, J.; Kucharczyk, R. Analysis of MT-ATP8 Gene Variants Reported in Patients by Modeling in Silico and in Yeast Model Organism. Sci. Rep. 2023, 13, 9972.
- Ware, S.M.; El-Hassan, N.; Kahler, S.G.; Zhang, Q.; Ma, Y.W.; Miller, E.; Wong, B.; Spicer, R.L.; Craigen, W.J.; Kozel, B.A.; et al. Infantile Cardiomyopathy Caused by a Mutation in the Overlapping Region of Mitochondrial ATPase 6 and 8 Genes. J. Med. Genet. 2009, 46, 308–314.
- Imai, A.; Fujita, S.; Kishita, Y.; Kohda, M.; Tokuzawa, Y.; Hirata, T.; Mizuno, Y.; Harashima, H.; Nakaya, A.; Sakata, Y.; et al. Rapidly Progressive Infantile Cardiomyopathy with Mitochondrial Respiratory Chain Complex V Deficiency Due to Loss of ATPase 6 and 8 Protein. Int. J. Cardiol. 2016, 207, 203–205.
- Jonckheere, A.I.; Hogeveen, M.; Nijtmans, L.G.J.; Van Den Brand, M.A.M.; Janssen, A.J.M.; Diepstra, J.H.S.; Van Den Brandt, F.C.A.; Van Den Heuvel, L.P.; Hol, F.A.; Hofste, T.G.J.; et al. A Novel Mitochondrial ATP8 Gene Mutation in a Patient with Apical Hypertrophic Cardiomyopathy and Neuropathy. J. Med. Genet. 2007, 45, 129–133.
- Boominathan, A.; Vanhoozer, S.; Basisty, N.; Powers, K.; Crampton, A.L.; Wang, X.; Friedricks, N.; Schilling, B.; Brand, M.D.; O’Connor, M.S. Stable Nuclear Expression of ATP8 and ATP6 Genes Rescues a mtDNA Complex V Null Mutant. Nucleic Acids Res. 2016, 44, 9342–9357.
- Kytövuori, L.; Lipponen, J.; Rusanen, H.; Komulainen, T.; Martikainen, M.H.; Majamaa, K. A Novel Mutation m.8561C>G in MT-ATP6/8 Causing a Mitochondrial Syndrome with Ataxia, Peripheral Neuropathy, Diabetes Mellitus, and Hypergonadotropic Hypogonadism. J. Neurol. 2016, 263, 2188–2195.
- Fragaki, K.; Chaussenot, A.; Serre, V.; Acquaviva, C.; Bannwarth, S.; Rouzier, C.; Chabrol, B.; Paquis-Flucklinger, V. A Novel Variant m.8561C>T in the Overlapping Region of MT-ATP6 and MT-ATP8 in a Child with Early-Onset Severe Neurological Signs. Mol. Genet. Metab. Rep. 2019, 21, 100543.
- Jackson, C.B.; Hahn, D.; Schröter, B.; Richter, U.; Battersby, B.J.; Schmitt-Mechelke, T.; Marttinen, P.; Nuoffer, J.-M.; Schaller, A. A Novel Mitochondrial ATP6 Frameshift Mutation Causing Isolated Complex V Deficiency, Ataxia and Encephalomyopathy. Eur. J. Med. Genet. 2017, 60, 345–351.
- Lopez-Gallardo, E.; Solano, A.; Herrero-Martin, M.D.; Martinez-Romero, I.; Castano-Perez, M.D.; Andreu, A.L.; Herrera, A.; Lopez-Perez, M.J.; Ruiz-Pesini, E.; Montoya, J. NARP Syndrome in a Patient Harbouring an Insertion in the MT-ATP6 Gene That Results in a Truncated Protein. J. Med. Genet. 2008, 46, 64–67.
- Bugiardini, E.; Bottani, E.; Marchet, S.; Poole, O.V.; Beninca, C.; Horga, A.; Woodward, C.; Lam, A.; Hargreaves, I.; Chalasani, A.; et al. Expanding the Molecular and Phenotypic Spectrum of Truncating MT-ATP6 Mutations. Neurol. Genet. 2020, 6, e381.
- Blanco-Grau, A.; Bonaventura-Ibars, I.; Coll-Cantí, J.; Melià, M.J.; Martinez, R.; Martínez-Gallo, M.; Andreu, A.L.; Pinós, T.; García-Arumí, E. Identification and Biochemical Characterization of the Novel Mutation m. 8839G>C in the Mitochondrial ATP6 Gene Associated with NARP Syndrome. Genes Brain Behav. 2013, 12, 812–820.
- Baranowska, E.; Niedzwiecka, K.; Panja, C.; Charles, C.; Dautant, A.; Poznanski, J.; Di Rago, J.-P.; Tribouillard-Tanvier, D.; Kucharczyk, R. Probing the Pathogenicity of Patient-Derived Variants of MT-ATP6 in Yeast. Dis. Models Mech. 2023, 16, dmm049783.
- Kucharczyk, R.; Giraud, M.-F.; Brèthes, D.; Wysocka-Kapcinska, M.; Ezkurdia, N.; Salin, B.; Velours, J.; Camougrand, N.; Haraux, F.; Di Rago, J.-P. Defining the Pathogenesis of Human mtDNA Mutations Using a Yeast Model: The Case of T8851C. Int. J. Biochem. Cell Biol. 2013, 45, 130–140.
- Ding, Q.; Kucharczyk, R.; Zhao, W.; Dautant, A.; Xu, S.; Niedzwiecka, K.; Su, X.; Giraud, M.-F.; Gombeau, K.; Zhang, M.; et al. Case Report: Identification of a Novel Variant (m.8909T>C) of Human Mitochondrial ATP6 Gene and Its Functional Consequences on Yeast ATP Synthase. Life 2020, 10, 215.
- Niedzwiecka, K.; Kabala, A.M.; Lasserre, J.-P.; Tribouillard-Tanvier, D.; Golik, P.; Dautant, A.; Di Rago, J.-P.; Kucharczyk, R. Yeast Models of Mutations in the Mitochondrial ATP6 Gene Found in Human Cancer Cells. Mitochondrion 2016, 29, 7–17.
- Abu-Amero, K.K.; Bosley, T.M. Detection of Mitochondrial Respiratory Dysfunction in Circulating Lymphocytes Using Resazurin. Arch. Pathol. Lab. Med. 2005, 129, 1295–1298.
- Wen, S.; Niedzwiecka, K.; Zhao, W.; Xu, S.; Liang, S.; Zhu, X.; Xie, H.; Tribouillard-Tanvier, D.; Giraud, M.-F.; Zeng, C.; et al. Identification of G8969>A in Mitochondrial ATP6 Gene That Severely Compromises ATP Synthase Function in a Patient with IgA Nephropathy. Sci. Rep. 2016, 6, 36313.
- Skoczeń, N.; Dautant, A.; Binko, K.; Godard, F.; Bouhier, M.; Su, X.; Lasserre, J.-P.; Giraud, M.-F.; Tribouillard-Tanvier, D.; Chen, H.; et al. Molecular Basis of Diseases Caused by the mtDNA Mutation m.8969G>A in the Subunit a of ATP Synthase. Biochim. Biophys. Acta (BBA)-Bioenerg. 2018, 1859, 602–611.
- Burrage, L.C.; Tang, S.; Wang, J.; Donti, T.R.; Walkiewicz, M.; Luchak, J.M.; Chen, L.-C.; Schmitt, E.S.; Niu, Z.; Erana, R.; et al. Mitochondrial Myopathy, Lactic Acidosis, and Sideroblastic Anemia (MLASA) plus Associated with a Novel de Novo Mutation (m.8969G>A) in the Mitochondrial Encoded ATP6 Gene. Mol. Genet. Metab. 2014, 113, 207–212.
- Duno, M.; Wibrand, F.; Baggesen, K.; Rosenberg, T.; Kjaer, N.; Frederiksen, A.L. A Novel Mitochondrial Mutation m.8989G>C Associated with Neuropathy, Ataxia, Retinitis Pigmentosa—The NARP Syndrome. Gene 2013, 515, 372–375.
- Rak, M.; Tetaud, E.; Duvezin-Caubet, S.; Ezkurdia, N.; Bietenhader, M.; Rytka, J.; Di Rago, J.-P. A Yeast Model of the Neurogenic Ataxia Retinitis Pigmentosa (NARP) T8993G Mutation in the Mitochondrial ATP Synthase-6 Gene. J. Biol. Chem. 2007, 282, 34039–34047.
- Baracca, A.; Barogi, S.; Carelli, V.; Lenaz, G.; Solaini, G. Catalytic Activities of Mitochondrial ATP Synthase in Patients with Mitochondrial DNA T8993G Mutation in the ATPase 6 Gene Encoding Subunit a. J. Biol. Chem. 2000, 275, 4177–4182.
- Carelli, V.; Baracca, A.; Barogi, S.; Pallotti, F.; Valentino, M.L.; Montagna, P.; Zeviani, M.; Pini, A.; Lenaz, G.; Baruzzi, A.; et al. Biochemical-Clinical Correlation in Patients with Different Loads of the Mitochondrial DNA T8993G Mutation. Arch. Neurol. 2002, 59, 264.
- Tatuch, Y.; Robinson, B.H. The Mitochondrial DNA Mutation at 8993 Associated with NARP Slows the Rate of ATP Synthesis in Isolated Lymphoblast Mitochondria. Biochem. Biophys. Res. Commun. 1993, 192, 124–128.
- Trounce, I.; Neill, S.; Wallace, D.C. Cytoplasmic Transfer of the mtDNA Nt 8993 T-->G (ATP6) Point Mutation Associated with Leigh Syndrome into mtDNA-Less Cells Demonstrates Cosegregation with a Decrease in State III Respiration and ADP/O Ratio. Proc. Natl. Acad. Sci. USA 1994, 91, 8334–8338.
- Sgarbi, G.; Baracca, A.; Lenaz, G.; Valentino, L.M.; Carelli, V.; Solaini, G. Inefficient Coupling between Proton Transport and ATP Synthesis May Be the Pathogenic Mechanism for NARP and Leigh Syndrome Resulting from the T8993G Mutation in mtDNA. Biochem. J. 2006, 395, 493–500.
- Baracca, A.; Sgarbi, G.; Mattiazzi, M.; Casalena, G.; Pagnotta, E.; Valentino, M.L.; Moggio, M.; Lenaz, G.; Carelli, V.; Solaini, G. Biochemical Phenotypes Associated with the Mitochondrial ATP6 Gene Mutations at Nt8993. Biochim. Biophys. Acta (BBA)-Bioenerg. 2007, 1767, 913–919.
- Solaini, G.; Harris, D.A.; Lenaz, G.; Sgarbi, G.; Baracca, A. The Study of the Pathogenic Mechanism of Mitochondrial Diseases Provides Information on Basic Bioenergetics. Biochim. Biophys. Acta (BBA)-Bioenerg. 2008, 1777, 941–945.
- Carrozzo, R.; Wittig, I.; Santorelli, F.M.; Bertini, E.; Hofmann, S.; Brandt, U.; Schägger, H. Subcomplexes of Human ATP Synthase Mark Mitochondrial Biosynthesis Disorders. Ann. Neurol. 2006, 59, 265–275.
- Vázquez-Memije, M.E.; Shanske, S.; Santorelli, F.M.; Kranz-Eble, P.; DeVivo, D.C.; DiMauro, S. Comparative Biochemical Studies of ATPases in Cells from Patients with the T8993G or T8993C Mitochondrial DNA Mutations. J. Inherit. Metab. Dis. 1998, 21, 829–836.
- Manfredi, G.; Gupta, N.; Vazquez-Memije, M.E.; Sadlock, J.E.; Spinazzola, A.; De Vivo, D.C.; Schon, E.A. Oligomycin Induces a Decrease in the Cellular Content of a Pathogenic Mutation in the Human Mitochondrial ATPase 6 Gene. J. Biol. Chem. 1999, 274, 9386–9391.
- García, J.J.; Ogilvie, I.; Robinson, B.H.; Capaldi, R.A. Structure, Functioning, and Assembly of the ATP Synthase in Cells from Patients with the T8993G Mitochondrial DNA Mutation. J. Biol. Chem. 2000, 275, 11075–11081.
- Cortés-Hernández, P.; Vázquez-Memije, M.E.; García, J.J. ATP6 Homoplasmic Mutations Inhibit and Destabilize the Human F1F0-ATP Synthase without Preventing Enzyme Assembly and Oligomerization. J. Biol. Chem. 2007, 282, 1051–1058.
- Bonnet, C.; Kaltimbacher, V.; Ellouze, S.; Augustin, S.; Bénit, P.; Forster, V.; Rustin, P.; Sahel, J.-A.; Corral-Debrinski, M. Allotopic mRNA Localization to the Mitochondrial Surface Rescues Respiratory Chain Defects in Fibroblasts Harboring Mitochondrial DNA Mutations Affecting Complex I or V Subunits. Rejuvenation Res. 2007, 10, 127–144.
- Lebiedzinska, M.; Karkucinska-Wieckowska, A.; Wojtala, A.; Suski, J.M.; Szabadkai, G.; Wilczynski, G.; Wlodarczyk, J.; Diogo, C.V.; Oliveira, P.J.; Tauber, J.; et al. Disrupted ATP Synthase Activity and Mitochondrial Hyperpolarisation-Dependent Oxidative Stress Is Associated with p66Shc Phosphorylation in Fibroblasts of NARP Patients. Int. J. Biochem. Cell Biol. 2013, 45, 141–150.
- Vazquez-Memije, M.E.; Rizza, T.; Meschini, M.C.; Nesti, C.; Santorelli, F.M.; Carrozzo, R. Cellular and Functional Analysis of Four Mutations Located in the Mitochondrial ATPase6 Gene. J Cell. Biochem. 2009, 106, 878–886.
- Mizuguchi, Y.; Hatakeyama, H.; Sueoka, K.; Tanaka, M.; Goto, Y. Low Dose Resveratrol Ameliorates Mitochondrial Respiratory Dysfunction and Enhances Cellular Reprogramming. Mitochondrion 2017, 34, 43–48.
- Uittenbogaard, M.; Brantner, C.A.; Fang, Z.; Wong, L.-J.C.; Gropman, A.; Chiaramello, A. Novel Insights into the Functional Metabolic Impact of an Apparent de Novo m.8993T>G Variant in the MT-ATP6 Gene Associated with Maternally Inherited Form of Leigh Syndrome. Mol. Genet. Metab. 2018, 124, 71–81.
- Geromel, V. Superoxide-Induced Massive Apoptosis in Cultured Skin Fibroblasts Harboring the Neurogenic Ataxia Retinitis Pigmentosa (NARP) Mutation in the ATPase-6 Gene of the Mitochondrial DNA. Hum. Mol. Genet. 2001, 10, 1221–1228.
- Nijtmans, L.G.J.; Henderson, N.S.; Attardi, G.; Holt, I.J. Impaired ATP Synthase Assembly Associated with a Mutation in the Human ATP Synthase Subunit 6 Gene. J. Biol. Chem. 2001, 276, 6755–6762.
- Manfredi, G.; Fu, J.; Ojaimi, J.; Sadlock, J.E.; Kwong, J.Q.; Guy, J.; Schon, E.A. Rescue of a Deficiency in ATP Synthesis by Transfer of MTATP6, a Mitochondrial DNA-Encoded Gene, to the Nucleus. Nat. Genet. 2002, 30, 394–399.
- Pallotti, F.; Baracca, A.; Hernandez-Rosa, E.; Walker, W.F.; Solaini, G.; Lenaz, G.; Melzi d’Eril, G.V.; Dimauro, S.; Schon, E.A.; Davidson, M.M. Biochemical Analysis of Respiratory Function in Cybrid Cell Lines Harbouring Mitochondrial DNA Mutations. Biochem. J. 2004, 384, 287–293.
- Mattiazzi, M. The mtDNA T8993G (NARP) Mutation Results in an Impairment of Oxidative Phosphorylation That Can Be Improved by Antioxidants. Hum. Mol. Genet. 2004, 13, 869–879.
- D’Aurelio, M.; Vives-Bauza, C.; Davidson, M.M.; Manfredi, G. Mitochondrial DNA Background Modifies the Bioenergetics of NARP/MILS ATP6 Mutant Cells. Hum. Mol. Genet. 2010, 19, 374–386.
- Carrozzo, R.; Rizza, T.; Lucioli, S.; Pierini, R.; Bertini, E.; Santorelli, F. A Mitochondrial ATPase 6 Mutation Is Associated with Leigh Syndrome in a Family and Affects Proton Flow and Adenosine Triphosphate Output When Modeled in Escherichia coli. Acta Paediatr. 2004, 93, 65–67.
- López-Gallardo, E.; Emperador, S.; Solano, A.; Llobet, L.; Martín-Navarro, A.; López-Pérez, M.J.; Briones, P.; Pineda, M.; Artuch, R.; Barraquer, E.; et al. Expanding the Clinical Phenotypes of MT-ATP6 Mutations. Hum. Mol. Genet. 2014, 23, 6191–6200.
- Gaude, E.; Schmidt, C.; Gammage, P.A.; Dugourd, A.; Blacker, T.; Chew, S.P.; Saez-Rodriguez, J.; O’Neill, J.S.; Szabadkai, G.; Minczuk, M.; et al. NADH Shuttling Couples Cytosolic Reductive Carboxylation of Glutamine with Glycolysis in Cells with Mitochondrial Dysfunction. Mol. Cell 2018, 69, 581–593.e7.
- Szczepanowska, J.; Zabłocki, K.; Duszyński, J. Influence of a Mitochondrial Genetic Defect on Capacitative Calcium Entry and Mitochondrial Organization in the Osteosarcoma Cells. FEBS Lett. 2004, 578, 316–322.
- Walczak, J.; Partyka, M.; Duszyński, J.; Szczepanowska, J. Implications of Mitochondrial Network Organization in Mitochondrial Stress Signalling in NARP Cybrid and Rho0 Cells. Sci. Rep. 2017, 7, 14864.
- Wojewoda, M.; Duszyński, J.; Szczepanowska, J. Antioxidant Defence Systems and Generation of Reactive Oxygen Species in Osteosarcoma Cells with Defective Mitochondria: Effect of Selenium. Biochim. Biophys. Acta (BBA)-Bioenerg. 2010, 1797, 890–896.
- Chen, Q.; Kirk, K.; Shurubor, Y.I.; Zhao, D.; Arreguin, A.J.; Shahi, I.; Valsecchi, F.; Primiano, G.; Calder, E.L.; Carelli, V.; et al. Rewiring of Glutamine Metabolism Is a Bioenergetic Adaptation of Human Cells with Mitochondrial DNA Mutations. Cell Metab. 2018, 27, 1007–1025.e5.
- Ma, H.; Folmes, C.D.L.; Wu, J.; Morey, R.; Mora-Castilla, S.; Ocampo, A.; Ma, L.; Poulton, J.; Wang, X.; Ahmed, R.; et al. Metabolic Rescue in Pluripotent Cells from Patients with mtDNA Disease. Nature 2015, 524, 234–238.
- Romero-Morales, A.I.; Robertson, G.L.; Rastogi, A.; Rasmussen, M.L.; Temuri, H.; McElroy, G.S.; Chakrabarty, R.P.; Hsu, L.; Almonacid, P.M.; Millis, B.A.; et al. Human iPSC-Derived Cerebral Organoids Model Features of Leigh Syndrome and Reveal Abnormal Corticogenesis. Development 2022, 149, dev199914.
- Zheng, X.; Boyer, L.; Jin, M.; Kim, Y.; Fan, W.; Bardy, C.; Berggren, T.; Evans, R.M.; Gage, F.H.; Hunter, T. Alleviation of Neuronal Energy Deficiency by mTOR Inhibition as a Treatment for Mitochondria-Related Neurodegeneration. eLife 2016, 5, e13378.
- Kucharczyk, R.; Rak, M.; Di Rago, J.-P. Biochemical Consequences in Yeast of the Human Mitochondrial DNA 8993T>C Mutation in the ATPase6 Gene Found in NARP/MILS Patients. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2009, 1793, 817–824.
- Sikorska, M.; Sandhu, J.K.; Simon, D.K.; Pathiraja, V.; Sodja, C.; Li, Y.; Ribecco-Lutkiewicz, M.; Lanthier, P.; Borowy-Borowski, H.; Upton, A.; et al. Identification of Ataxia-associated mtDNA Mutations (m.4052T>C and m.9035T>C) and Evaluation of Their Pathogenicity in Transmitochondrial Cybrids. Muscle Nerve 2009, 40, 381–394.
- Capiau, S.; Smet, J.; De Paepe, B.; Yildiz, Y.; Arslan, M.; Stevens, O.; Verschoore, M.; Stepman, H.; Seneca, S.; Vanlander, A. Clinical Heterogeneity in MT-ATP6 Pathogenic Variants: Same Genotype—Different Onset. Cells 2022, 11, 489.
- Lamminen, T.; Majander, A.; Juvonen, V.; Wikström, M.; Aula, P.; Nikoskelainen, E.; Savontous, M.L. A Mitochondrial Mutation at Nt 9101 in the ATP Synthase 6 Gene Associated with Deficient Oxidative Phosphorylation in a Family with Leber Hereditary Optic Neuroretinopathy. Am. J. Hum. Genet. 1995, 56, 1238–1240.
- Majander, A.; Lamminen, T.; Juvonen, V.; Aula, P.; Nikoskelainen, E.; Savontaus, M.-L.; Wikström, M. Mutations in Subunit 6 of the F1F0-ATP Synthase Cause Two Entirely Different Diseases. FEBS Lett. 1997, 412, 351–354.
- Mordel, P.; Schaeffer, S.; Dupas, Q.; Laville, M.-A.; Gérard, M.; Chapon, F.; Allouche, S. A 2 Bp Deletion in the Mitochondrial ATP 6 Gene Responsible for the NARP (Neuropathy, Ataxia, and Retinitis Pigmentosa) Syndrome. Biochem. Biophys. Res. Commun. 2017, 494, 133–137.
- Honzik, T.; Tesarova, M.; Magner, M.; Mayr, J.; Jesina, P.; Vesela, K.; Wenchich, L.; Szentivanyi, K.; Hansikova, H.; Sperl, W.; et al. Neonatal Onset of Mitochondrial Disorders in 129 Patients: Clinical and Laboratory Characteristics and a New Approach to Diagnosis. J. Inherit. Metab. Dis. 2012, 35, 749–759.
- Kenvin, S.; Torregrosa-Muñumer, R.; Reidelbach, M.; Pennonen, J.; Turkia, J.J.; Rannila, E.; Kvist, J.; Sainio, M.T.; Huber, N.; Herukka, S.-K.; et al. Threshold of Heteroplasmic Truncating MT-ATP6 Mutation in Reprogramming, Notch Hyperactivation and Motor Neuron Metabolism. Hum. Mol. Genet. 2022, 31, 958–974.
- Kucharczyk, R.; Salin, B.; Di Rago, J.-P. Introducing the Human Leigh Syndrome Mutation T9176G into Saccharomyces Cerevisiae Mitochondrial DNA Leads to Severe Defects in the Incorporation of Atp6p into the ATP Synthase and in the Mitochondrial Morphology. Hum. Mol. Genet. 2009, 18, 2889–2898.
- Carrozzo, R.; Tessa, A.; Vazquez-Memije, M.E.; Piemonte, F.; Patrono, C.; Malandrini, A.; Dionisi-Vici, C.; Vilarinho, L.; Villanova, M.; Schagger, H.; et al. The T9176G mtDNA Mutation Severely Affects ATP Production and Results in Leigh Syndrome. Neurology 2001, 56, 687–690.
- Kucharczyk, R.; Ezkurdia, N.; Couplan, E.; Procaccio, V.; Ackerman, S.H.; Blondel, M.; Di Rago, J.-P. Consequences of the Pathogenic T9176C Mutation of Human Mitochondrial DNA on Yeast Mitochondrial ATP Synthase. Biochim. Biophys. Acta (BBA)-Bioenerg. 2010, 1797, 1105–1112.
- Thyagarajan, D.; Shanske, S.; Vazquez -Memije, M.; Devivo, D.; Dimauro, S. A Novel Mitochondrial ATPase 6 Point Mutation in Familial Bilateral Striatal Necrosis. Ann. Neurol. 1995, 38, 468–472.
- Verny, C.; Guegen, N.; Desquiret, V.; Chevrollier, A.; Prundean, A.; Dubas, F.; Cassereau, J.; Ferre, M.; Amati-Bonneau, P.; Bonneau, D.; et al. Hereditary Spastic Paraplegia-like Disorder Due to a Mitochondrial ATP6 Gene Point Mutation. Mitochondrion 2011, 11, 70–75.
- Kabala, A.M.; Lasserre, J.-P.; Ackerman, S.H.; Di Rago, J.-P.; Kucharczyk, R. Defining the Impact on Yeast ATP Synthase of Two Pathogenic Human Mitochondrial DNA Mutations, T9185C and T9191C. Biochimie 2014, 100, 200–206.
- Moslemi, A.-R.; Darin, N.; Tulinius, M.; Oldfors, A.; Holme, E. Two New Mutations in the MTATP6 Gene Associated with Leigh Syndrome. Neuropediatrics 2005, 36, 314–318.
- Pitceathly, R.D.S.; Murphy, S.M.; Cottenie, E.; Chalasani, A.; Sweeney, M.G.; Woodward, C.; Mudanohwo, E.E.; Hargreaves, I.; Heales, S.; Land, J.; et al. Genetic Dysfunction of MT-ATP6 Causes Axonal Charcot-Marie-Tooth Disease. Neurology 2012, 79, 1145–1154.
- Castagna, A.E.; Addis, J.; McInnes, R.R.; Clarke, J.T.R.; Ashby, P.; Blaser, S.; Robinson, B.H. Late Onset Leigh Syndrome and Ataxia Due to a T to C Mutation at Bp 9,185 of Mitochondrial DNA. Am. J. Med. Genet. 2007, 143A, 808–816.
- Saneto, R.P.; Singh, K.K. Illness-Induced Exacerbation of Leigh Syndrome in a Patient with the MTATP6 Mutation, m. 9185 T>C. Mitochondrion 2010, 10, 567–572.
- Pfeffer, G.; Blakely, E.L.; Alston, C.L.; Hassani, A.; Boggild, M.; Horvath, R.; Samuels, D.C.; Taylor, R.W.; Chinnery, P.F. Adult-Onset Spinocerebellar Ataxia Syndromes Due to MTATP6 Mutations. J. Neurol. Neurosurg. Psychiatry 2012, 83, 883–886.
- Lorenz, C.; Lesimple, P.; Bukowiecki, R.; Zink, A.; Inak, G.; Mlody, B.; Singh, M.; Semtner, M.; Mah, N.; Auré, K.; et al. Human iPSC-Derived Neural Progenitors Are an Effective Drug Discovery Model for Neurological mtDNA Disorders. Cell Stem Cell 2017, 20, 659–674.e9.
- Kucharczyk, R.; Dautant, A.; Gombeau, K.; Godard, F.; Tribouillard-Tanvier, D.; Di Rago, J.-P. The Pathogenic MT-ATP6 m.8851T>C Mutation Prevents Proton Movements within the n-Side Hydrophilic Cleft of the Membrane Domain of ATP Synthase. Biochim. Biophys. Acta (BBA)-Bioenerg. 2019, 1860, 562–572.
- Ješina, P.; Tesařová, M.; Fornůsková, D.; Vojtíšková, A.; Pecina, P.; Kaplanová, V.; Hansíková, H.; Zeman, J.; Houštěk, J. Diminished Synthesis of Subunit a (ATP6) and Altered Function of ATP Synthase and Cytochrome c Oxidase Due to the mtDNA 2 Bp Microdeletion of TA at Positions 9205 and 9206. Biochem. J. 2004, 383, 561–571.
- Seneca, S.; Abramowicz, M.; Lissens, W.; Muller, M.F.; Vamos, E.; De Meirleir, L. A Mitochondrial DNA Microdeletion in a Newborn Girl with Transient Lactic Acidosis. J. Inherit. Metab. Dis. 1996, 19, 115–118.
- Dautant, A.; Meier, T.; Hahn, A.; Tribouillard-Tanvier, D.; Di Rago, J.-P.; Kucharczyk, R. ATP Synthase Diseases of Mitochondrial Genetic Origin. Front. Physiol. 2018, 9, 329.
- Jonckheere, A.I.; Smeitink, J.A.M.; Rodenburg, R.J.T. Mitochondrial ATP Synthase: Architecture, Function and Pathology. J. Inherit. Metab. Dis. 2012, 35, 211–225.
- Ganetzky, R.D.; Stendel, C.; McCormick, E.M.; Zolkipli-Cunningham, Z.; Goldstein, A.C.; Klopstock, T.; Falk, M.J. MT-ATP6 Mitochondrial Disease Variants: Phenotypic and Biochemical Features Analysis in 218 Published Cases and Cohort of 14 New Cases. Hum. Mutat. 2019, 40, 499–515.
- Morava, E.; Rodenburg, R.J.; Hol, F.; De Vries, M.; Janssen, A.; Van Den Heuvel, L.; Nijtmans, L.; Smeitink, J. Clinical and Biochemical Characteristics in Patients with a High Mutant Load of the Mitochondrial T8993G/C Mutations. Am. J. Med. Genet. 2006, 140A, 863–868.
- Na, J.; Lee, Y. Genotype-phenotype Analysis of MT-ATP6 -associated Leigh Syndrome. Acta Neuro Scand. 2022, 145, 414–422.
- Solaini, G.; Sgarbi, G.; Lenaz, G.; Baracca, A. Evaluating Mitochondrial Membrane Potential in Cells. Biosci. Rep. 2007, 27, 11–21.
- Rieger, B.; Arroum, T.; Borowski, M.-T.; Villalta, J.; Busch, K.B. Mitochondrial F1FO ATP Synthase Determines the Local Proton Motive Force at Cristae Rims. EMBO Rep. 2021, 22, e52727.
- Spikes, T.E.; Montgomery, M.G.; Walker, J.E. Interface Mobility between Monomers in Dimeric Bovine ATP Synthase Participates in the Ultrastructure of Inner Mitochondrial Membranes. Proc. Natl. Acad. Sci. USA 2021, 118, e2021012118.
- Bénit, P.; El-Khoury, R.; Schiff, M.; Sainsard-Chanet, A.; Rustin, P. Genetic Background Influences Mitochondrial Function: Modeling Mitochondrial Disease for Therapeutic Development. Trends Mol. Med. 2010, 16, 210–217.
- Wilkins, H.M.; Carl, S.M.; Swerdlow, R.H. Cytoplasmic Hybrid (Cybrid) Cell Lines as a Practical Model for Mitochondriopathies. Redox Biol. 2014, 2, 619–631.
- McKnight, C.L.; Low, Y.C.; Elliott, D.A.; Thorburn, D.R.; Frazier, A.E. Modelling Mitochondrial Disease in Human Pluripotent Stem Cells: What Have We Learned? IJMS 2021, 22, 7730.
- Palombo, F.; Peron, C.; Caporali, L.; Iannielli, A.; Maresca, A.; Di Meo, I.; Fiorini, C.; Segnali, A.; Sciacca, F.L.; Rizzo, A.; et al. The Relevance of Mitochondrial DNA Variants Fluctuation during Reprogramming and Neuronal Differentiation of Human iPSCs. Stem Cell Rep. 2021, 16, 1953–1967.
- Lorenz, C.; Zink, A.; Henke, M.-T.; Staege, S.; Mlody, B.; Bünning, M.; Wanker, E.; Diecke, S.; Schuelke, M.; Prigione, A. Generation of Four iPSC Lines from Four Patients with Leigh Syndrome Carrying Homoplasmic Mutations m.8993T > G or m.8993T > C in the Mitochondrial Gene MT-ATP6. Stem Cell Res. 2022, 61, 102742.
- Galera-Monge, T.; Zurita-Díaz, F.; Moreno-Izquierdo, A.; Fraga, M.F.; Fernández, A.F.; Ayuso, C.; Garesse, R.; Gallardo, M.E. Generation of a Human iPSC Line from a Patient with an Optic Atrophy “plus” Phenotype Due to a Mutation in the OPA1 Gene. Stem Cell Res. 2016, 16, 673–676.
- Grace, H.E.; Galdun, P.; Lesnefsky, E.J.; West, F.D.; Iyer, S. mRNA Reprogramming of T8993G Leigh’s Syndrome Fibroblast Cells to Create Induced Pluripotent Stem Cell Models for Mitochondrial Disorders. Stem Cells Dev. 2019, 28, 846–859.
- Henke, M.-T.; Zink, A.; Diecke, S.; Prigione, A.; Schuelke, M. Generation of Two Mother–Child Pairs of iPSCs from Maternally Inherited Leigh Syndrome Patients with m.8993 T > G and m.9176 T > G MT-ATP6 Mutations. Stem Cell Res. 2023, 67, 103030.
- Lai, Y.; Zhang, Y.; Zhou, S.; Xu, J.; Du, Z.; Feng, Z.; Yu, L.; Zhao, Z.; Wang, W.; Tang, Y.; et al. Structure of the Human ATP Synthase. Mol. Cell 2023, 83, 2137–2147.e4.
- Steiner, T.; Zink, A.; Henke, M.-T.; Cecchetto, G.; Buenning, M.; Rossi, A.; Schuelke, M.; Prigione, A. RNA-Based Generation of iPSCs from a Boy Carrying the Mutation m.9185 T>C in the Mitochondrial Gene MT-ATP6 and from His Healthy Mother. Stem Cell Res. 2022, 64, 102920.
- Licchetta, L.; Ferri, L.; La Morgia, C.; Zenesini, C.; Caporali, L.; Lucia Valentino, M.; Minardi, R.; Fulitano, D.; Di Vito, L.; Mostacci, B.; et al. Epilepsy in MT - ATP6 - Related Mils/NARP: Correlation of Elettroclinical Features with Heteroplasmy. Ann. Clin. Transl. Neurol. 2021, 8, 704–710.
- Angural, A.; Sharma, I.; Pandoh, P.; Sharma, V.; Spolia, A.; Rai, E.; Singh, V.; Razdan, S.; Pandita, K.K.; Sharma, S. A Case Report on a Novel MT-ATP6 Gene Variation in Atypical Mitochondrial Leigh Syndrome Associated with Bilateral Basal Ganglia Calcifications. Mitochondrion 2019, 46, 209–213.
- Lehmann Urban, D.; Motlagh Scholle, L.; Wagner, M.; Ludolph, A.C.; Rosenbohm, A. The m.9143T>C Variant: Recurrent Infections and Immunodeficiency as an Extension of the Phenotypic Spectrum in MT-ATP6 Mutations? Diseases 2020, 8, 19.
- Sainio, M.T.; Aaltio, J.; Hyttinen, V.; Kortelainen, M.; Ojanen, S.; Paetau, A.; Tienari, P.; Ylikallio, E.; Auranen, M.; Tyynismaa, H. Effectiveness of Clinical Exome Sequencing in Adult Patients with Difficult-to-diagnose Neurological Disorders. Acta Neuro Scand. 2022, 145, 63–72.
- Birtel, J.; Von Landenberg, C.; Gliem, M.; Gliem, C.; Reimann, J.; Kunz, W.S.; Herrmann, P.; Betz, C.; Caswell, R.; Nesbitt, V.; et al. Mitochondrial Retinopathy. Ophthalmol. Retin. 2022, 6, 65–79.
- Nolte, D.; Kang, J.-S.; Hofmann, A.; Schwaab, E.; Krämer, H.H.; Müller, U. Mutations in MT-ATP6 Are a Frequent Cause of Adult-Onset Spinocerebellar Ataxia. J. Neurol. 2021, 268, 4866–4873.
