Non-coding RNAs (ncRNAs) are RNA molecules that do not code for protein but play key roles in regulating cellular processes. NcRNAs globally affect gene expression in diverse physiological and pathological contexts. Functionally important ncRNAs act in chromatin modifications, mRNA stabilization and translation, and the regulation of various signaling pathways. Non-alcoholic fatty liver disease (NAFLD) is a set of conditions caused by the accumulation of triacylglycerol in the liver. Studies of ncRNA in NAFLD are limited but have demonstrated that ncRNAs play a critical role in the pathogenesis of NAFLD.
- ncRNA
- miRNA
- PiwiRNA
- LncRNA
- CircRNA
- NAFLD
- NASH
- oxidative stress
- metabolic dysfunction
- insulin resistance
1. Introduction
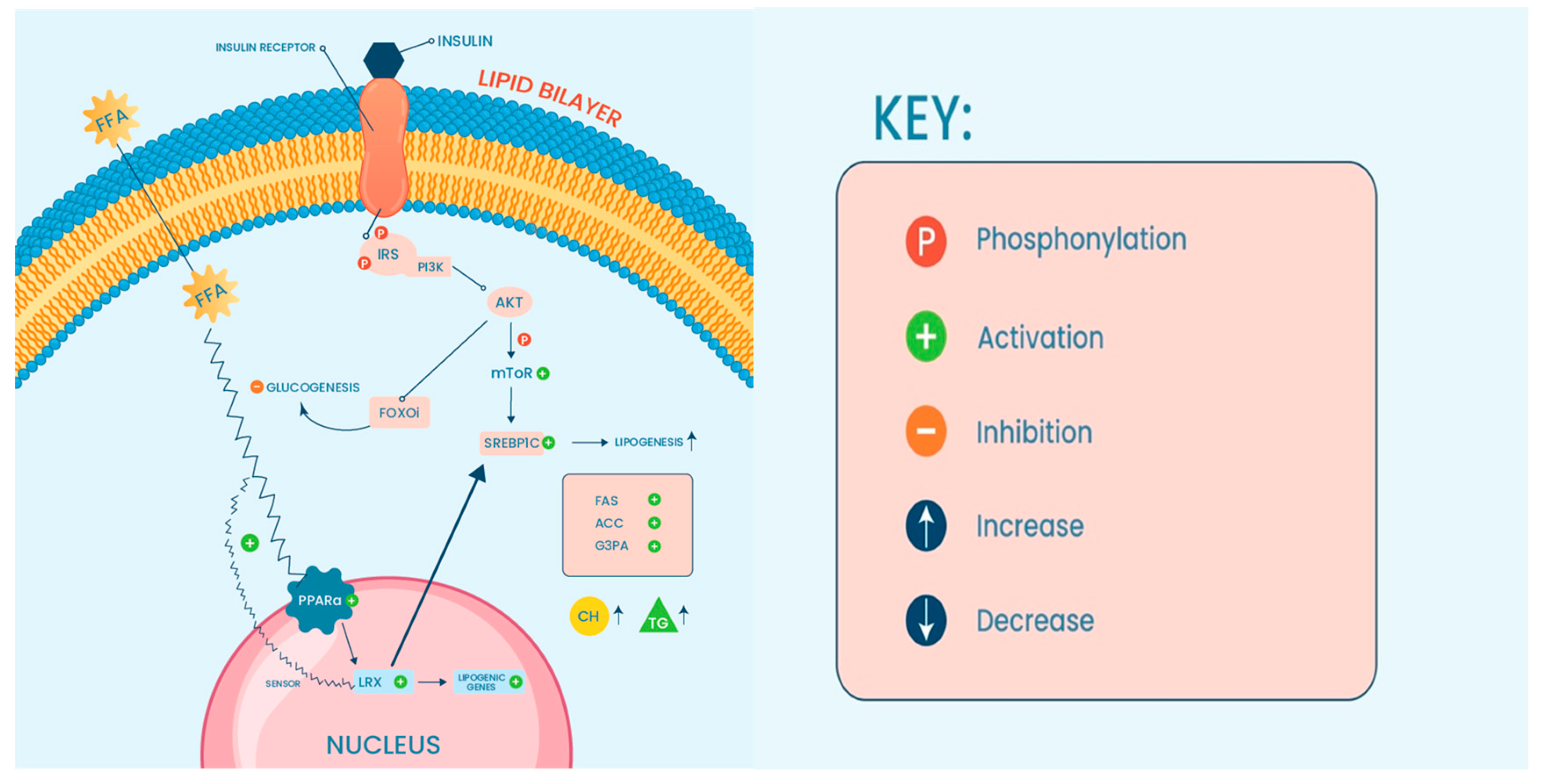
2. Liver Cellular Function and Development of NAFLD/NASH
3. NcRNA and Intercellular Communication in Liver Cell Populations
4. NcRNA and the Dysregulated Pathways Related to NAFLD/NASH
|
Non-Coding RNA |
Expression |
Primary Function |
Reference |
|---|---|---|---|
|
miR-96-124-182-183 |
Upregulated |
These ncRNAs modulate lipid synthesis through SREBP-1c |
|
|
miR-206 |
Downregulated |
[22] |
|
|
Gm16551 |
Downregulated |
[23] |
|
|
lncHR1 |
Downregulated |
[24] |
|
|
miR-122-3075-21-192-5p |
Upregulated |
These ncRNAs are involved in the stimulation or inhibition of HCs and HC-related injuries |
|
|
piR-823 |
Upregulated |
[34] |
|
|
circBNC2 |
Upregulated |
[35] |
|
|
circ_0071410 |
[32] |
||
|
circPI4KB |
[31] |
||
|
circRNA_0046367 |
Downregulated |
[36] |
|
|
miR-26 |
Downregulated |
miR-26 expression depends on LXRs and has a protective role |
[37] |
|
miR-34a |
Upregulated |
Both ncRNAs were found to be associated with the regulation of hepatic fat content through PPARα |
[38] |
|
circRNA_0046366 |
Downregulated |
[38] |
|
|
miR-451 |
Downregulated |
Involved regulation of glucose homeostasis |
[39] |
|
miR-103/miR-107 |
Upregulated |
[40] |
|
|
LncARSR |
Upregulated |
The expression of LncARSR correlates with the activated state of PI3K/AKT/mTOR pathway |
[41] |
|
miR-155 |
Upregulated |
These ncRNASs have an influence on the induction of inflammation via NF-κB pathway |
|
|
Platr4 |
Upregulated |
[44] |
|
|
LncTNF |
Upregulated |
[45] |
|
|
miR-125 |
Upregulated |
The expression of these ncRNAs contributes to the activation of pro-inflammatory mediators |
[46] |
|
circFBXW4 |
Downregulated |
[14] |
|
|
miR-29b |
Downregulated |
These ncRNAs are involved in fibrosis mediation through TGF-β1 |
[33] |
|
H19 |
Downregulated |
||
|
piR-823 |
Upregulated |
[34] |
|
|
miR-26a-143-145 |
Upregulated |
These ncRNAs have been found to be modulators in insulin sensitivity |
|
|
AK085787-uc009kuu.1-uc008txr.1 |
Upregulated |
[23] |
|
|
CircRNF111-circRNA_0001805 |
Downregulated |
||
|
CircRNA SCAR |
Upregulated |
This ncRNA may play a role in activating liver fibroblasts |
[54] |
5. The Role of ncRNA in NAFLD-Associated Metabolic Dysfunction
This entry is adapted from the peer-reviewed paper 10.3390/ncrna10010010
References
- Huang, R.; Duan, X.; Fan, J.; Li, G.; Wang, B. Role of Noncoding RNA in Development of Nonalcoholic Fatty Liver Disease. Biomed. Res. Int. 2019, 2019, 8690592.
- Riazi, K.; Azhari, H.; Charette, J.H.; Underwood, F.E.; King, J.A.; Afshar, E.E.; Swain, M.G.; Congly, S.E.; Kaplan, G.G.; Shaheen, A.A. The prevalence and incidence of NAFLD worldwide: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2022, 7, 851–861.
- Del Campo, J.A.; Gallego-Durán, R.; Gallego, P.; Grande, L. Genetic and Epigenetic Regulation in Nonalcoholic Fatty Liver Disease (NAFLD). Int. J. Mol. Sci. 2018, 19, 911.
- Jirtle, R.L.; Skinner, M.K. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 2007, 8, 253–262.
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055.
- Peterson, C.L.; Workman, J.L. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000, 10, 187–192.
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610.
- Litwin, M.; Szczepańska-Buda, A.; Piotrowska, A.; Dzięgiel, P.; Witkiewicz, W. The meaning of PIWI proteins in cancer development. Oncol. Lett. 2017, 13, 3354–3362.
- Han, Y.N.; Li, Y.; Xia, S.Q.; Zhang, Y.Y.; Zheng, J.H.; Li, W. PIWI Proteins and PIWI-Interacting RNA: Emerging Roles in Cancer. Cell. Physiol. Biochem. 2017, 44, 1–20.
- Trefts, E.; Gannon, M.; Wasserman, D.H. The liver. Curr. Biol. 2017, 27, R1147–r1151.
- Gong, J.; Tu, W.; Liu, J.; Tian, D. Hepatocytes: A key role in liver inflammation. Front. Immunol. 2022, 13, 1083780.
- Legaki, A.I.; Moustakas, I.I.; Sikorska, M.; Papadopoulos, G.; Velliou, R.I.; Chatzigeorgiou, A. Hepatocyte Mitochondrial Dynamics and Bioenergetics in Obesity-Related Non-Alcoholic Fatty Liver Disease. Curr. Obes. Rep. 2022, 11, 126–143.
- Malhi, H.; Guicciardi, M.E.; Gores, G.J. Hepatocyte death: A clear and present danger. Physiol. Rev. 2010, 90, 1165–1194.
- Chen, X.; Li, H.D.; Bu, F.T.; Li, X.F.; Chen, Y.; Zhu, S.; Wang, J.N.; Chen, S.Y.; Sun, Y.Y.; Pan, X.Y.; et al. Circular RNA circFBXW4 suppresses hepatic fibrosis via targeting the miR-18b-3p/FBXW7 axis. Theranostics 2020, 10, 4851–4870.
- Chen, J.; Deng, X.; Liu, Y.; Tan, Q.; Huang, G.; Che, Q.; Guo, J.; Su, Z. Kupffer Cells in Non-alcoholic Fatty Liver Disease: Friend or Foe? Int. J. Biol. Sci. 2020, 16, 2367–2378.
- Miyao, M.; Kotani, H.; Ishida, T.; Kawai, C.; Manabe, S.; Abiru, H.; Tamaki, K. Pivotal role of liver sinusoidal endothelial cells in NAFLD/NASH progression. Lab. Investig. 2015, 95, 1130–1144.
- Wiering, L.; Subramanian, P.; Hammerich, L. Hepatic Stellate Cells: Dictating Outcome in Nonalcoholic Fatty Liver Disease. Cell. Mol. Gastroenterol. Hepatol. 2023, 15, 1277–1292.
- Stout, R.D.; Suttles, J. Functional plasticity of macrophages: Reversible adaptation to changing microenvironments. J. Leukoc. Biol. 2004, 76, 509–513.
- Zeng, Q.; Liu, C.H.; Wu, D.; Jiang, W.; Zhang, N.; Tang, H. LncRNA and circRNA in Patients with Non-Alcoholic Fatty Liver Disease: A Systematic Review. Biomolecules 2023, 13, 560.
- Jeon, T.I.; Esquejo, R.M.; Roqueta-Rivera, M.; Phelan, P.E.; Moon, Y.A.; Govindarajan, S.S.; Esau, C.C.; Osborne, T.F. An SREBP-responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis. Cell Metab. 2013, 18, 51–61.
- Liu, D.D.; Han, C.C.; Wan, H.F.; He, F.; Xu, H.Y.; Wei, S.H.; Du, X.H.; Xu, F. Effects of inhibiting PI3K-Akt-mTOR pathway on lipid metabolism homeostasis in goose primary hepatocytes. Animal 2016, 10, 1319–1327.
- Wu, H.; Zhang, T.; Pan, F.; Steer, C.J.; Li, Z.; Chen, X.; Song, G. MicroRNA-206 prevents hepatosteatosis and hyperglycemia by facilitating insulin signaling and impairing lipogenesis. J. Hepatol. 2017, 66, 816–824.
- Yang, L.; Li, P.; Yang, W.; Ruan, X.; Kiesewetter, K.; Zhu, J.; Cao, H. Integrative Transcriptome Analyses of Metabolic Responses in Mice Define Pivotal LncRNA Metabolic Regulators. Cell Metab. 2016, 24, 627–639.
- Li, D.; Guo, L.; Deng, B.; Li, M.; Yang, T.; Yang, F.; Yang, Z. Long non-coding RNA HR1 participates in the expression of SREBP-1c through phosphorylation of the PDK1/AKT/FoxO1 pathway. Mol. Med. Rep. 2018, 18, 2850–2856.
- Ji, Y.; Luo, Z.; Gao, H.; Dos Reis, F.C.G.; Bandyopadhyay, G.; Jin, Z.; Manda, K.A.; Isaac, R.; Yang, M.; Fu, W.; et al. Hepatocyte-derived exosomes from early onset obese mice promote insulin sensitivity through miR-3075. Nat. Metab. 2021, 3, 1163–1174.
- Pourhoseini, S.; Seth, R.K.; Das, S.; Dattaroy, D.; Kadiiska, M.B.; Xie, G.; Michelotti, G.A.; Nagarkatti, M.; Diehl, A.M.; Chatterjee, S. Upregulation of miR21 and repression of Grhl3 by leptin mediates sinusoidal endothelial injury in experimental nonalcoholic steatohepatitis. PLoS ONE 2015, 10, e0116780.
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98.
- Pirola, C.J.; Fernández Gianotti, T.; Castaño, G.O.; Mallardi, P.; San Martino, J.; Mora Gonzalez Lopez Ledesma, M.; Flichman, D.; Mirshahi, F.; Sanyal, A.J.; Sookoian, S. Circulating microRNA signature in non-alcoholic fatty liver disease: From serum non-coding RNAs to liver histology and disease pathogenesis. Gut 2015, 64, 800–812.
- Inomata, Y.; Oh, J.W.; Taniguchi, K.; Sugito, N.; Kawaguchi, N.; Hirokawa, F.; Lee, S.W.; Akao, Y.; Takai, S.; Kim, K.P.; et al. Downregulation of miR-122-5p Activates Glycolysis via PKM2 in Kupffer Cells of Rat and Mouse Models of Non-Alcoholic Steatohepatitis. Int. J. Mol. Sci. 2022, 23, 5230.
- Das, A.; Basu, S.; Bandyopadhyay, D.; Mukherjee, K.; Datta, D.; Chakraborty, S.; Jana, S.; Adak, M.; Bose, S.; Chakrabarti, S.; et al. Inhibition of extracellular vesicle-associated MMP2 abrogates intercellular hepatic miR-122 transfer to liver macrophages and curtails inflammation. iScience 2021, 24, 103428.
- Liu, C.H.; Jiang, W.; Zeng, Q.; Wu, D.; Li, H.; Zhou, L.; Bai, L.; Tang, H. CircRNA-PI4KB Induces Hepatic Lipid Deposition in Non-Alcoholic Fatty Liver Disease by Transporting miRNA-122 to Extra-Hepatocytes. Int. J. Mol. Sci. 2023, 24, 1297.
- Chien, Y.; Tsai, P.H.; Lai, Y.H.; Lu, K.H.; Liu, C.Y.; Lin, H.F.; Huang, C.S.; Wu, W.W.; Wang, C.Y. CircularRNA as novel biomarkers in liver diseases. J. Chin. Med. Assoc. 2020, 83, 15–17.
- Dattaroy, D.; Pourhoseini, S.; Das, S.; Alhasson, F.; Seth, R.K.; Nagarkatti, M.; Michelotti, G.A.; Diehl, A.M.; Chatterjee, S. Micro-RNA 21 inhibition of SMAD7 enhances fibrogenesis via leptin-mediated NADPH oxidase in experimental and human nonalcoholic steatohepatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G298–G312.
- Tang, X.; Xie, X.; Wang, X.; Wang, Y.; Jiang, X.; Jiang, H. The Combination of piR-823 and Eukaryotic Initiation Factor 3 B (EIF3B) Activates Hepatic Stellate Cells via Upregulating TGF-β1 in Liver Fibrogenesis. Med. Sci. Monit. 2018, 24, 9151–9165.
- Wang, P.; Huang, Z.; Peng, Y.; Li, H.; Lin, T.; Zhao, Y.; Hu, Z.; Zhou, Z.; Zhou, W.; Liu, Y.; et al. Circular RNA circBNC2 inhibits epithelial cell G2-M arrest to prevent fibrotic maladaptive repair. Nat. Commun. 2022, 13, 6502.
- Guo, X.Y.; Chen, J.N.; Sun, F.; Wang, Y.Q.; Pan, Q.; Fan, J.G. circRNA_0046367 Prevents Hepatoxicity of Lipid Peroxidation: An Inhibitory Role against Hepatic Steatosis. Oxid. Med. Cell. Longev. 2017, 2017, 3960197.
- Sun, D.; Zhang, J.; Xie, J.; Wei, W.; Chen, M.; Zhao, X. MiR-26 controls LXR-dependent cholesterol efflux by targeting ABCA1 and ARL7. FEBS Lett. 2012, 586, 1472–1479.
- Guo, X.Y.; Sun, F.; Chen, J.N.; Wang, Y.Q.; Pan, Q.; Fan, J.G. circRNA_0046366 inhibits hepatocellular steatosis by normalization of PPAR signaling. World J. Gastroenterol. 2018, 24, 323–337.
- Zhuo, S.; Yang, M.; Zhao, Y.; Chen, X.; Zhang, F.; Li, N.; Yao, P.; Zhu, T.; Mei, H.; Wang, S.; et al. MicroRNA-451 Negatively Regulates Hepatic Glucose Production and Glucose Homeostasis by Targeting Glycerol Kinase-Mediated Gluconeogenesis. Diabetes 2016, 65, 3276–3288.
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653.
- Huang, J.; Chen, S.; Cai, D.; Bian, D.; Wang, F. Long noncoding RNA lncARSR promotes hepatic cholesterol biosynthesis via modulating Akt/SREBP-2/HMGCR pathway. Life Sci. 2018, 203, 48–53.
- Shen, M.; Pan, H.; Ke, J.; Zhao, F. NF-κB-upregulated miR-155-5p promotes hepatocyte mitochondrial dysfunction to accelerate the development of nonalcoholic fatty liver disease through downregulation of STC1. J. Biochem. Mol. Toxicol. 2022, 36, e23025.
- Tili, E.; Michaille, J.J.; Cimino, A.; Costinean, S.; Dumitru, C.D.; Adair, B.; Fabbri, M.; Alder, H.; Liu, C.G.; Calin, G.A.; et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007, 179, 5082–5089.
- Lin, Y.; Wang, S.; Gao, L.; Zhou, Z.; Yang, Z.; Lin, J.; Ren, S.; Xing, H.; Wu, B. Oscillating lncRNA Platr4 regulates NLRP3 inflammasome to ameliorate nonalcoholic steatohepatitis in mice. Theranostics 2021, 11, 426–444.
- Shi, N.; Sun, K.; Tang, H.; Mao, J. The impact and role of identified long noncoding RNAs in nonalcoholic fatty liver disease: A narrative review. J. Clin. Lab. Anal. 2023, 37, e24943.
- Zhang, Q.; Yu, K.; Cao, Y.; Luo, Y.; Liu, Y.; Zhao, C. miR-125b promotes the NF-κB-mediated inflammatory response in NAFLD via directly targeting TNFAIP3. Life Sci. 2021, 270, 119071.
- Li, X.; Liu, R.; Wang, Y.; Zhu, W.; Zhao, D.; Wang, X.; Yang, H.; Gurley, E.C.; Chen, W.; Hylemon, P.B.; et al. Cholangiocyte-Derived Exosomal lncRNA H19 Promotes Macrophage Activation and Hepatic Inflammation under Cholestatic Conditions. Cells 2020, 9, 190.
- Jiang, X.; Ning, Q. The mechanism of lncRNA H19 in fibrosis and its potential as novel therapeutic target. Mech. Ageing Dev. 2020, 188, 111243.
- Fu, X.; Dong, B.; Tian, Y.; Lefebvre, P.; Meng, Z.; Wang, X.; Pattou, F.; Han, W.; Wang, X.; Lou, F.; et al. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. J. Clin. Investig. 2015, 125, 2497–2509.
- Xihua, L.; Shengjie, T.; Weiwei, G.; Matro, E.; Tingting, T.; Lin, L.; Fang, W.; Jiaqiang, Z.; Fenping, Z.; Hong, L. Circulating miR-143-3p inhibition protects against insulin resistance in Metabolic Syndrome via targeting of the insulin-like growth factor 2 receptor. Transl. Res. 2019, 205, 33–43.
- Jordan, S.D.; Krüger, M.; Willmes, D.M.; Redemann, N.; Wunderlich, F.T.; Brönneke, H.S.; Merkwirth, C.; Kashkar, H.; Olkkonen, V.M.; Böttger, T.; et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat. Cell Biol. 2011, 13, 434–446.
- Li, J.; Qi, J.; Tang, Y.; Liu, H.; Zhou, K.; Dai, Z.; Yuan, L.; Sun, C. A nanodrug system overexpressed circRNA_0001805 alleviates nonalcoholic fatty liver disease via miR-106a-5p/miR-320a and ABCA1/CPT1 axis. J. Nanobiotechnology 2021, 19, 363.
- Lin, X.; Du, Y.; Lu, W.; Gui, W.; Sun, S.; Zhu, Y.; Wang, G.; Eserberg, D.T.; Zheng, F.; Zhou, J.; et al. CircRNF111 Protects Against Insulin Resistance and Lipid Deposition via Regulating miR-143-3p/IGF2R Axis in Metabolic Syndrome. Front. Cell Dev. Biol. 2021, 9, 663148.
- Zhao, Q.; Liu, J.; Deng, H.; Ma, R.; Liao, J.Y.; Liang, H.; Hu, J.; Li, J.; Guo, Z.; Cai, J.; et al. Targeting Mitochondria-Located circRNA SCAR Alleviates NASH via Reducing mROS Output. Cell 2020, 183, 76–93.
- Méndez-Sánchez, N.; Díaz-Orozco, L.E. Editorial: International Consensus Recommendations to Replace the Terminology of Non-Alcoholic Fatty Liver Disease (NAFLD) with Metabolic-Associated Fatty Liver Disease (MAFLD). Med. Sci. Monit. 2021, 27, e933860.
- Dietrich, P.; Hellerbrand, C. Non-alcoholic fatty liver disease, obesity and the metabolic syndrome. Best. Pract. Res. Clin. Gastroenterol. 2014, 28, 637–653.
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84.
- Godoy-Matos, A.F.; Silva Júnior, W.S.; Valerio, C.M. NAFLD as a continuum: From obesity to metabolic syndrome and diabetes. Diabetol. Metab. Syndr. 2020, 12, 60.
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Charlton, M.; Cusi, K.; Rinella, M.; Harrison, S.A.; Brunt, E.M.; Sanyal, A.J. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018, 67, 328–357.
- Czech, M.P. Insulin action and resistance in obesity and type 2 diabetes. Nat. Med. 2017, 23, 804–814.
