1. Introduction
Zinc finger and BTB domain-containing (ZBTB) proteins are an evolutionarily conserved family of transcription factors (TF), characterized by the presence of an N-terminal BTB domain and one or more C-terminal Cys2His2 (C2H2)/Krüppel-type zinc finger (ZF) domains [
1]. So far, at least 49 ZBTB proteins have been identified in the human genome (
Figure 1), exhibiting diverse functions in both physiological and pathological conditions [
1]. Of these members, ZBTB20 was initially identified as a dendritic cell (DC)-derived ZBTB protein (also named DPZF, HOF, or ZNF288) [
2], acting mainly as a transcription repressor involved in many biological processes, including cellular differentiation, developmental regulation, innate immunity, and metabolism [
3,
4,
5,
6,
7,
8,
9]. ZBTB20 has also been increasingly identified as a key regulator in the pathogenesis and development of human cancers.
Figure 1. Schematic representation of ZBTB family members and their encoded domains. The sequences of ZBTB proteins and their functional information are obtained from the UniProk Knowledgebase (UniProtKB) [
10] and National Center for Biotechnology Information (NCBI). aa, amino acids; BCL6, BCL6 transcription repressor (also known as ZBTB27); BCL6B, also known as ZBTB28; BTB, bric-a-brac/tramtrack/broad complex; C2H2-type ZF, C-terminal Cys2His2/Krüppel-type zinc finger; chr, chromosome; GZF1, GDNF-inducible zinc finger protein 1 (also known as ZBTB23); HIC1, HIC ZBTB transcriptional repressor 1 (also known as ZBTB29); HIC2, also known as ZBTB30; MYNN, myoneurin (also known as ZBTB31); NLS, nuclear localization signal; PATZ1, POZ/BTB and AT hook-containing zinc finger 1 (also known as ZBTB19); ZNF131, zinc finger protein 131 (also known as ZBTB35).
2. Functional Domains of ZBTB Proteins
2.1. The BTB Domain
The BTB domain, also known as the POZ (poxvirus and zinc finger) domain, is a conserved protein–protein interaction motif widely distributed in the proteins of higher eukaryotes [
11]. It has approximately 100–120 amino acids (aa) within the N terminus of ZBTB protein (
Figure 1). Despite being structurally well conserved [
12], the BTB domain participates in a variety of cellular processes. It mediates not only the formation of homodimers, heterodimers, or multimers [
12,
13,
14], but also the recruitment of transcriptional regulators [
15,
16], hence allowing for diverse functions, ranging from transcriptional repression [
17,
18,
19] and cytoskeleton dynamics [
20] to protein ubiquitination and degradation [
21,
22,
23,
24]. BTB domains are also capable of regulating gene expression through chromatin remodeling [
25]. Given its essential roles, the BTB domain has become a promising target for some diseases. Numerous studies have demonstrated the successful pharmacological inhibition of the BTB domain by small-molecule and peptide-mimicking compounds [
26,
27,
28,
29,
30,
31], although its therapeutic use needs further evaluation.
2.2. The C2H2-Type ZF Domain
The C2H2-type ZF domains are one of the most common DNA-binding motifs found in the TFs of higher eukaryotes [
32]. The classical C2H2 domain has 25–30 aa with a left-handed ββα structure, which consists of a β-hairpin followed by an α-helix. Although C2H2 domains contain highly conserved cysteine and histidine pairs, the other aa residues are variable [
32], and the number of C2H2-type ZF domains in each ZBTB protein varies too (
Figure 1). These variations enable ZBTB proteins to have a unique DNA-binding capability in a sequence-specific manner [
33,
34]. Moreover, C2H2-type ZFs also mediate protein–protein interactions. For example, the ZFs of ZBTB16 (also known as PLZF) mediates its interaction with GATA-binding protein 2 (GATA2) [
35]. The association between ZBTB16 and the histone acetyltransferase E1A-binding protein p300 (EP300) also depends on the acetylation of lysines in ZFs of ZBTB16 [
36].
2.3. Other Known Domains
In addition to BTB and ZF domains, further sequence analyses have revealed other motifs or domains. So far, ZBTB14, ZBTB33, myoneurin (MYNN, also known as ZBTB31), and zinc finger protein 131 (ZNF131, also known as ZBTB35) have been identified to have one or two nuclear localization signals (NLS) (
Figure 1), which are short peptides that act as a signal fragment to mediate the transport of proteins from the cytoplasm into the nucleus [
37]. In addition, ZBTB24 and POZ/BTB and AT hook-containing zinc finger 1 (PATZ1, also known as ZBTB19) contain an A-T hook (
Figure 1), a DNA-binding domain that interacts with the minor groove of AT-rich regions, thus facilitating the binding of other TFs [
38,
39]. Specifically, the A-T hook domain of ZBTB24 is essential for heterochromatin localization [
40], whereas the region containing the A-T hook of PATZ1 is sufficient to interact with ring finger protein 4 (RNF4), a transcriptional repressor [
41].
2.4. The Linker Region
In nearly all ZBTB proteins, the BTB and ZF domains are connected by a long linker region, which is predicted to be unstructured in most cases and confers flexible DNA-binding properties [
12]. However, very limited studies have been carried out in this field. Based on the existing research, the linker region appears to act as a key mediator with specific effects on the regulation of DNA-binding capacity and protein stability [
1].
2.5. ZBTB Family Members
In humans, the ZBTB family comprises a large group of TFs, many of which serve as master regulators of developmental events. Among them, BCL6 transcription repressor (BCL6), also known as ZBTB27, is a hallmark member controlling germinal center (GC) formation and T follicular helper cell differentiation [
42,
43,
44,
45,
46]. By contrast, ZBTB16 directs the function and development of natural killer T (NKT) cells [
47,
48]; ZBTB20 promotes plasma cell differentiation but restricts T-cell memory differentiation [
7,
9]; ZBTB32 is essential for the proliferation of NK cells and limits the duration of antibody recall responses [
49,
50]. Due to their pivotal roles in hematopoiesis and immune responses, it is not surprising that abnormal expression or dysfunction of ZBTB proteins leads to hematological malignancies. Indeed, the first characterized mammalian ZBTB protein was from the study of chromosomal translocations in human acute promyelocytic leukemia (APL). Analysis of the t (11;17) translocation from a case with APL led to the discovery of
ZBTB16, which is fused to the retinoic acid receptor alpha (
RARA), resulting in the worst prognosis in APL [
51]. Another example is the deregulated BCL6 expression which not only causes B-cell lymphoma development [
52] but also contributes to leukemia initiation [
53,
54]. Consequently, BCL6 has become an attractive therapeutic target for B-cell lymphomas and leukemias [
54]. In addition to hematological malignancies, ZBTB family proteins are also involved in skeletal abnormalities [
55,
56], infertility [
57,
58,
59], and neurological disorders [
60,
61,
62].
Of ZBTB family members,
ZBTB20 encodes a 741-residue protein with an N-terminal BTB domain and 5 C2H2-type ZF domains at the C terminus [
63]. Notably,
ZBTB20 is localized on the chromosome 3 where
BCL6 is located and shares high homology to
BCL6 with the identity of 56% in the BTB domain and 40% in the C2H2-type ZF domain; particularly, ZBTB20 is widely expressed in hematopoietic tissues [
2]. These similarities imply that ZBTB20 may be highly close to BCL6 with a certain role in hematopoiesis, immune responses, and oncogenesis.
3. Physiological Roles of ZBTB20
3.1. Lymphoid Development and Differentiation
ZBTB20 was originally identified in human DCs and widely expressed in hematopoietic tissues including the spleen, lymph node, thymus, peripheral blood cells, and fetal liver [
2]. In mice, Zbtb20 is highly expressed in B1 and GC B cells and reaches its highest level in mature plasma cells in the bone marrow (BM) [
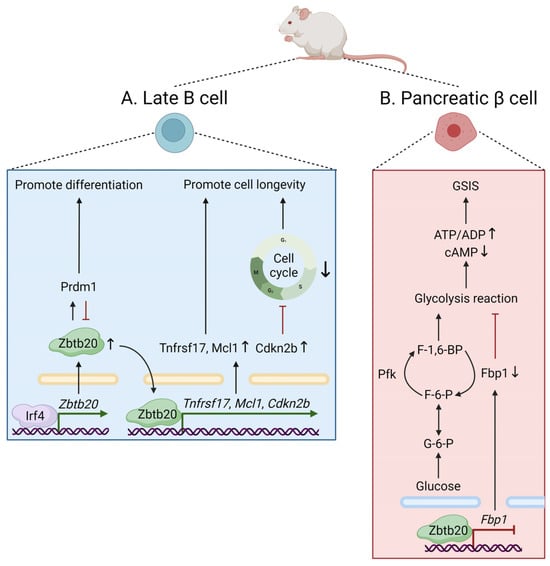
7]. Of note, ectopic expression of Zbtb20 in primary B cells facilitates plasma cell differentiation, whereas in plasma cells, Zbtb20 promotes cell longevity by inducing cell survival and cell cycle arrest (
Figure 2A) [
7]. These roles are opposite to those played by Bcl6, reflecting a possible competition between Zbtb20 and Bcl6 for binding to common gene targets. Moreover, Zbtb20 is also required for long-term antibody production and plasma cell persistence, specifically after alum-adjuvanted immunization [
8], revealing Zbtb20 as a potential molecular determinant to drive durable antibody response.
Figure 2. The physiological roles of Zbtb20 in mice. (A) Zbtb20 facilitates plasma cell differentiation via an Irf4/Zbtb20/Prdm1 axis, in which Zbtb20 induction does not require Prdm1 but depends on Irf4 by directly binding to the Zbtb20 promoter. Zbtb20 also induces cell survival and blocks cell cycle progression by regulating the Tnfrsf17, Mcl1, and Cdkn2b genes, thus promoting plasma cell longevity. (B) In murine pancreatic β cells, Zbtb20 regulates glucose sensing and insulin secretion by repressing Fbp1, which encodes a gluconeogenic enzyme that regulates glucose metabolism and insulin secretion in β cells. ADP, adenosine diphosphate; ATP, adenosine triphosphate; cAMP, cyclic adenosine monophosphate; Cdkn2b, cyclin-dependent kinase inhibitor 2B; F-1,6-BP, fructose-1,6-bisphosphate; F-6-P, fructose-6-phosphate; Fbp1, fructose-1,6-bisphosphatase 1; G-6-P, glucose-6-phosphate; GSIS, glucose-stimulated insulin secretion; Irf4, interferon regulatory factor 4; Mcl1, myeloid cell leukemia sequence 1; Pfk, phosphofructokinase. Prdm1, PR domain-containing 1, with ZNF domain; Tnfrsf17, tumor necrosis factor receptor superfamily, member 17.
Apart from lymphoid differentiation, ZBTB20 is also involved in the functional roles of regulatory B/T (Breg/Treg) cells. In mice,
Zbtb20 is upregulated in Breg cells from all organs, suggesting that
Zbtbt20 may serve as a potential marker gene for Breg cells [
65]. By contrast, Zbtb20 expression defines the function of a subset of Treg cells involved in intestinal integrity [
66]. Specifically, nearly half of Zbtb20-expressing (Zbtb20
+) T cells express forkhead box protein P3 (FoxP3), a lineage-defining transcription factor for Treg cells. These Zbtb20
+ Treg cells are enriched in the intestine and constitutively transcribe interleukin 10 (
IL10), a key cytokine for modulating intestinal homeostasis. As such, Zbtb20 conditional knockout mice display severe intestinal inflammation and damage in response to induction of acute colitis, whereas adoptive transfer of Zbtb20
+ Treg cells, but not non-Zbtb20 Tregs, rescues the mice from colitis. However, apart from the intestine, Zbtb20
+ Treg cells with a similar ability to produce IL10 are also detected in the thymus and spleen, indicating that Zbtb20
+ Tregs are a committed population rather than an induced effector.
3.2. Cellular Metabolism
In addition to T-cell immunometabolism mediated by Zbtb20, as mentioned [
9], multiple lines of evidence have revealed the function of ZBTB20 in the regulation of cellular metabolism. In
Zbtb20-null mice, transcriptional profiling of liver tissue reveals significant alterations in the expression of genes involved in glucose metabolism [
3]. Further analysis revealed that Zbtb20 is abundantly expressed in pancreatic β cells and plays a key role in regulating glucose sensing and insulin secretion via transcriptional repression of fructose-1,6-bisphosphatase 1 (
Fbp1), a regulator of glucose metabolism and insulin secretion in β cells [
5] (
Figure 2B). In hepatocytes, Zbtb20 also regulates plasma triglyceride metabolism by repressing the transcriptional activity of lipoprotein lipase, highlighting a critical role of ZBTB20 for hepatic lipogenesis in mice [
67]. Such metabolic disorders mediated by ZBTB20 become more evident in patients with Primrose syndrome, an autosomal dominant disease caused by heterozygous missense variants in
ZBTB20, manifested by multisystem failures including disturbed lipid and glucose metabolism as well as mitochondrial dysfunction [
68,
69].
3.3. Neurodevelopment
In central nervous system (CNS), Zbtb20 was initially characterized in hippocampal neurons, cerebellar granule neurons, and macroglia, and two Zbtb20 isoforms, designated
Zbtb20(S) and
Zbtb20(L), were identified [
63]. Ectopic expression of Zbtb20(S) and/or Zbtb20(L) in non-hippocampal immature pyramidal neurons induces hippocampus (Hi)-like cortical neurogenesis in the mouse brain, but similar phenotypes are observed in Zbtb20(S), Zbtb20(L), and Zbtb20(S/L) transgenic mice, suggesting an overlap in function of these two isoforms [
70]. Furthermore, Zbtb20 has a dynamic expression pattern in the germinative zones of the developing neocortex and serves as a regulator of the timed sequential production of distinct neuronal fates during cortical neurogenesis [
71].
Specifically, Zbtb20 is essential for the specification of hippocampal Cornu Ammonis 1 (CA1) field identity and the postnatal survival of hippocampal neurons [
62]. The mice with specific deletion of
Zbtb20 in mature CA1 pyramidal neurons exhibit no obvious morphological abnormalities but display impaired Hi-dependent memory, demonstrating that Zbtb20 is critical for both the specification of CA1 field identity in the developing Hi and Hi-dependent long-term memory formation in mature CA1 cells [
72]. Subsequent studies further uncovered that Zbtb20 is highly expressed in all the mature endocrine cell types of anterior pituitary, and its deficiency impairs anterior pituitary development.
Zbtb20-null mice also display severe defects in lactotrope specification and lineage expansion, pinpointing that Zbtb20 is a crucial determinant of lactotrope specification [
73]. Recently, the same group dissected the roles of Zbtb20 in mature lactotropes.
3.4. Immune Response and Inflammation
Being a master regulator of lymphoid development and differentiation, ZBTB20 plays a key role in immune response and inflammation. For instance, Zbtb20 promotes full activation of Toll-like receptor (TLR) signaling, a critical pathway in innate response against invading pathogens; mechanistically, Zbtb20 enhances NF-κB activation by specifically repressing the transcription of
Nfkbia (
IκBα), the canonical suppressor of NF-κB signaling, thus promoting TLR-triggered production of proinflammatory cytokines and type I interferon (IFN) in macrophages [
6]. Consistent with this study, ZBTB20 affects the outcome of chlamydia and salmonella infections through the suppression of multiple target genes including
NFKBIA [
75]. ZBTB20 has also been revealed to regulate cardiac allograft rejection via macrophage polarization and NF-κB-mediated inflammation [
76]. Other than ZBTB20, circular RNA (circRNA)
circZbtb20 has been newly identified to promote the homeostasis and function of group 3 innate lymphoid cells (ILC3s), a group of innate effectors involved in host defense against pathogens at the early stage [
77]. All these findings pinpoint that ZBTB20 is a pleotropic regulator of immune response and inflammation.
4. ZBTB20 in Cancers
Despite the involvement of ZBTB20 in various cellular processes under healthy conditions, understanding of the roles of ZBTB20 in malignancies is still in its infancy. Over the past decades, ZBTB20 has been well researched in liver cancer even though most elegant investigations were limited to mice. In addition, many ZBTB20 variants have been identified in some cancers, but the pathogenic mechanisms are poorly understood. In this case, we summarize the known mechanisms and potential functions of ZBTB20 in different cancers (Table 1).
Table 1. Overview of ZBTB20 roles in cancers.
|
Cancer Type
|
Potential Function/Mechanism
|
Ref.
|
|
HCC
|
Mice
|
Transcriptional repression of the Afp gene
|
[78,79]
|
|
(1) Promotion of liver regeneration
(2) Regulation of the hepatic expression of Egfr
|
[80,117]
|
|
One of nineteen highly significant candidate locus implicated in mouse HCC
|
[82]
|
|
Humans
|
Reactivation of AFP via the miR-122-mediated regulation
|
[83]
|
|
An independent marker for poor prognosis in human HCC
|
[84,85]
|
|
Promotion of HCC by suppressing FOXO1
|
[85]
|
|
Promotion of HCC by correlation with SETD7
|
[86]
|
|
Suppression of HCV infection
|
[75]
|
|
Association with HBV integration frequency
|
[87]
|
|
GC
|
Humans
|
Identification of rs9841504 as a new susceptibility locus for non-cardia GC in the Chinese population
|
[88]
|
|
Association of rs9841504 with severe intestinal metaplasia/atypical hyperplasia in the Chinese population
|
[89]
|
|
Identification of rs758277701 in the MSI subtype of GC in the Korean population
|
[92]
|
|
Identification of rs9288999 as a protective factor for reducing GC risk in the Chinese Han population
|
[93]
|
|
Promotion of GC via the NFKBIA/NF-κB signaling pathway
|
[94]
|
|
CNS cancer
|
Humans
|
Glioma
|
A mutation hotspot
|
[95,96]
|
|
Neuronal and mixed neuronal-glial tumors
|
Implication of miRNAs that target ZBTB20 in the classification of pediatric cases
|
[97]
|
|
Low-grade glioma
|
Identification of ZBTB20-AS4 as a critical IncRNA for predicting the prognosis
|
[98,99]
|
|
Shh-MB
|
Identification of the fusion transcripts of ZBTB20 as recurrent fusions
|
[100]
|
|
Glioblastoma
|
Promotion of glioblastoma through the miR-758-5p/ZBTB20 axis or by ZBTB20
|
[101]
|
|
Blood cancer
|
Humans
|
B-CLL
|
The top differentially expressed gene in terms of VH mutation status
|
[102]
|
|
AML
|
Promotion of cell growth and migration via the LINC00641/miR-378a/ZBTB20 axis
|
[103]
|
|
Promotion of malignant phenotypes via the circ-SFMBT2/miR-582-3p/ZBTB20 axis
|
[104]
|
|
Promotion of leukemia development via the circ-0001602/miR-192-5p/ZBTB20 axis
|
[105]
|
|
MCL
|
(1) A novel downstream target repressed by BACH1
(2) Involvement in the BACH1-mediated regulation of tumor immune microenvironment
|
[106]
|
|
Others
|
Humans
|
BC
|
Downregulation in ERα+ BC biopsies upon treatment of aromatase inhibitors
|
[109]
|
|
Upregulation in ERα+ BC cell lines upon anacardic acid treatment
|
[110]
|
|
Promotion of cell migration and invasion via the SNHG8/miR-634/ZBTB20 axis
|
[111]
|
|
Promotion of cell proliferation, migration, and invasion via the circ-0104345/miR-876-3p/ZBTB20 axis
|
[112]
|
|
Colorectal cancer
|
Identification of rs10511330 and rs16822593 as two of the top 10 SNPs in patients
|
[113]
|
|
Cervical cancer
|
One of ten potential driver genes
|
[114]
|
|
NSCLC
|
(1) Upregulation in NSCLC tissues
(2) Promotion of cell proliferation by repressing FOXO1
|
[115]
|
|
Ovarian cancer
|
Increase in cells that migrate in omentum tissue pretreated with extracellular vesicles isolated from ascitic supernatant of high-grade patients
|
[116]
|
Note: AML, acute myeloid leukemia; AFP/Afp, alpha-fetoprotein; BACH1, BTB and CNC homology 1; BC, breast cancer; B-CLL, B-cell chronic lymphoblastic leukemia; circ-SFMBT2, circular RNA Scm-like with four mbt domains 2; Egfr, epithelial growth factor receptor; ERα+, estrogen receptor α positive; FOXO1, forkhead box O1; GC, gastric cancer; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; IncRNA, long non-coding RNA; miRNA, microRNA; MCL, mantle cell lymphoma; MSI, microsatellite instability; NSCLC, non-small cell lung cancer; Ref., references; SETD7, SET domain-containing 7; Shh-MB, Sonic Hedgehog medulloblastoma; SNHG8, small nucleolus RNA host gene 8; SNPs, single nucleotide polymorphisms.
This entry is adapted from the peer-reviewed paper 10.3390/biom14020192