Cardiovascular diseases are the leading cause of death worldwide, and arterial hypertension is a recognized cardiovascular risk factor that is responsible for high morbidity and mortality. Arterial hypertension is the result of an inflammatory process that results in the remodeling and thickening of the vascular walls, which is associated with an immunological response. Previous studies have attempted to demonstrate the relationship between oral disease, inflammation, and the development of systemic diseases. The existence of an association between periodontitis and hypertension is a controversial issue because the underlying pathophysiological processes and inflammatory mechanisms common to both diseases are unknown.
- arterial hypertension
- periodontitis
- inflammation
- miRNAs
1. Introduction
2. Hypertension
3. Hypertension and Inflammation
4. Inflammation and Periodontitis
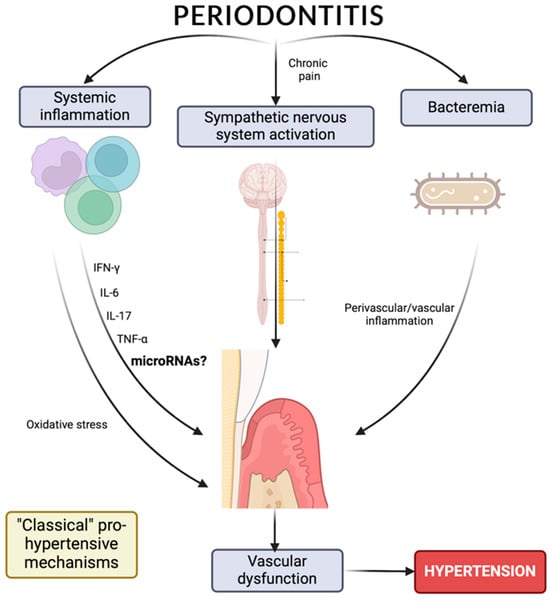
5. Hypertension, Periodontitis, and Inflammation
6. MicroRNAs as Risk Biomarkers for Hypertension and/or Periodontitis
| MicroRNAs | Model | Sample Size | Main Finding | Reference |
|---|---|---|---|---|
| miR-33a-5p, -144-3p | In vivo | Blood samples from 84 unrelated subjects (42 patients diagnosed with arterial hypertension and 42 normotensive subjects) | Hypertensive patients vs. control group: increased expression of miR-33a (p = 0.001) and miR-144 (p = 0.985), decreased expression of ABCA1 (p = 0.007) and ABCG1 (p = 0.550) transporters. | [76] |
| miR-153 | In vivo | Arteries from male normotensive Wistar rats and spontaneously hypertensive rats ranging from 12 to 16 in age | Increased expression of miR 153 in the arteries of spontaneously hypertensive rats (SHR) that exhibited decreased levels of Kv7.4. | [20] |
| miR-145 | In vivo and in vitro | Thoracic aortas from 10 26-week-old male spontaneously hypertensive rats and 10 age-matched normotensive male Wistar–Kyoto rats as control group Rat vascular endothelial cells isolated from the thoracic aortas |
miR-145 functions as a key mediator in the pathogenesis of hypertension through SLC7A1 targeting. | [77] |
| miR-126 | In vitro | HUVEC cells | Endothelial cells express miR-126, which inhibits VCAM-1 expression. | [78] |
| miR-146 | In vitro | THP-1, U937, HL-60, WEHI-3, 293/IL-1R/MD2/TLR4, BJAB, and Mono-Mac-6 cells | The role of miR-146 in the control of toll-like receptor and cytokine signaling through the downregulation of IL-1 kinase 1 and TNF receptor-associated factor 6 protein levels. | [79] |
| miR-296-5p, -let-7e, hcmv-miR-UL112. | In vivo | Whole blood from 194 hypertensive patients and 97 healthy volunteers | Twenty-seven differentially expressed miRNAs. Expressions of miR-296-5p, let-7e, and a human cytomegalovirus (HCMV)-encoded miRNA, hcmv-miR-UL112 were validated in plasma samples from 24 hypertensive patients and 22 control subjects. | [80] |
| miR-637 | In vitro | PC12 rat pheochromocytoma cells | ATP6V0A1 expression was affected by differential effects of miR-637, altering vacuolar pH and consequently CHGA processing and exocytotic secretion. | [81] |
| miR-1 | In vivo | Hearts isolated from Wistar rats divided into six groups: control, isoproterenol, ischaemia, ischaemia–propranolol, ischaemia–propranolol-miR-1, and ischaemia–AMO (n = 7 for each group) | The beta-adrenergic pathway can stimulate arrhythmogenic miR-1 expression, contributing to ischemic arrhythmogenesis, and beta-blockers produce their beneficial effects in part by downregulating miR-1. | [82] |
| miR-483-3p | In vivo and in vitro | Hearts obtained from age and gender matched transgenic mice over-expressing the human AT1R under the mouse α-MHC promoter and corresponding littermate non-transgenic mouse lines in C3H and C57BL/6 genetic backgrounds (n = 3 for each for group) Primary human aortic smooth muscle cells |
AT1R-regulated expression levels of angiotensin-1 and angiotensin-1 converting enzyme (ACE-1) proteins in VSMCs are specifically modulated by miR-483-3p. | [83] |
| miR-221, -222, -155 | In vitro | HUVEC cells | Increased expression of miR-155 and miR-221 in HUVEC and VSMC cells. The angiotensin II type 1 receptor (AT1R) is a target of miR-155 in HUVEC. Ets-1 and its downstream genes, including VCAM1, MCP1, and FLT1, were upregulated in angiotensin II-stimulated HUVECs, and this effect was partially reversed by overexpression of miR-155 and miR-221/2 | [84] |
| miR-208, -155 | In vivo | Aortas from 8-, 16-, and 24-week-old spontaneously hypertensive rats and 8-, 16-, and 24-week-old normotensive male Wistar–Kyoto rats as control group | The miR-155 level was negatively correlated with blood pressure (r = −0.525, p < 0.05). The expression of miR-208 in the aorta of hypertensive rats was negatively correlated with blood pressure (r = −0.400, p < 0.05) and age (r = −0.684, p < 0.0001). | [85] |
| MiR-21, -122, -637, -let-7e | In vivo | Plasma samples from 30 hypertension patients, 30 white coat hypertension patients, and 30 normotensive subjects | The expression levels of MiR-21, miR-122, miR-637, and let-7e were significantly increased in the group of hypertensive subjects compared to the group of normotensive subjects (p = 0.017, p = 0.022, p = 0.048 and p = 0.013, respectively). | [86] |
| miR-34a, -21, -126, -146a | In vivo | Plasma samples from 15 normotensive and 15 hypertensive subjects | Circulating expression of miR-34a was higher (~170%; p < 0.01) whereas expression of miR-21, miR-126, and miR-146a were markedly lower (~50%, ~55%, and ~55% respectively; p < 0.05) in the hypertensive versus normotensive. | [87] |
| miR-212, -132 | In vivo | Internal mammary artery with AngII receptor blockers (n = 16) and β-blockers (n = 9) from patients undergoing coronary artery by-pass graft surgery | miR-132 and miR-212 were upregulated in the heart, aortic wall, and kidney of rats with hypertension (159 ± 12 mm Hg) and cardiac hypertrophy after chronic Ang II infusion. In addition, activation of the endothelin receptor, another Gαq-coupled receptor, also increased miR-132 and miR-212. | [88] |
| miR-208b, -133a | In vivo | Blood and urine samples from 102 subjects with untreated newly diagnosed essential hypertension | miRNA-208b and miRNA-133a showed distinct profiles in peripheral blood cells isolated from untreated patients with newly diagnosed hypertension. Their gene expression levels revealed a strong correlation with urinary albumin excretion levels. | [89] |
| miR-25, -29a, -26b | In vivo | Blood samples from 104 acute Stanford type A aortic dissection + patients (of which 74 with hypertension and 30 without hypertension), and 103 age-matched acute Stanford type A aortic dissection individuals (of which 59 with hypertension and 44 without hypertension | 4-miRNA (miR-25, miR-29a, and miR-155) were significantly elevated, while miR-26b was decreased in AAAD+ serum samples compared to AAAD individuals), which may serve as a non-invasive biomarker for the diagnosis of AAAD, especially for subjects with hypertension. | [90] |
| miR-505 | In vivo and in vitro | Peripheral blood from 101 hypertensive patients and 91 healthy volunteers HUVEC cells |
The plasma level of hsa-miR-505 was significantly elevated in hypertensive patients. | [91] |
| miR-92a | In vivo | Plasma samples from 60 healthy volunteers with normal carotid intima-media thickness (nCIMT), 60 healthy volunteers with increased CIMT (iCIMT), 60 hypertensive patients with nCIMT and 60 hypertensive patients with iCIMT | miR-92a levels showed a significant positive correlation with mean 24-h systolic blood pressure (r = 0.807, p < 0.001), mean 24-h diastolic blood pressure (r = 0.649, p < 0.001), pulse pressure 24-h mean (PP) (r = 0.697, p < 0.001), 24-h daytime PP (r = 0.654, p < 0.001), 24-h nighttime PP (r = 0.573, p < 0.001), CIMT (r = 0.571, p < 0.001) and cfPWV (r = 0.601, p < 0.001). | [92] |
| miR-29a, -29b, -29c | In vivo | Whole blood from 54 patients with untreated hypertension and 30 healthy individuals | It was observed higher expression levels of miR-29a (p < 0.001), miR-29b (p < 0.001), and miR-29c (p < 0.001) in hypertensive patients compared to healthy control individuals. | [93] |
| miR-30e, -374b, -21 | - | Data obtained from Gene Expression Omnibus database | It was identified three crucial genes in the hypertensive kidney, such as COL12A1, ASPN, and SCN2A. ASPN could work in conjunction with COL12A1, and both could be targets for miR-21. SCN2A could be a new target for miR-30e and miR-374b. | [94] |
| miR-9, -126 | In vivo | Peripheral blood mononuclear cells of patients with essential hypertension (n = 60) and of healthy controls (n = 29) for comparison | Hypertensive patients showed significantly lower miR-9 (p < 0.001) and miR-126 (p < 0.001) expression levels compared to healthy controls. | [95] |
| miR-425, -155 | In vitro | Human embryonic stem cell-derived cardiomyocytes | The combination of miR-425 and miR-155 reduced NPPA expression to a greater extent than miR-425 or miR-155 alone, regardless of whether they separately also reduced NPPA expression. | [96] |
| miR-223 | In vivo | Samples from lean (n = 19) and obese (n = 19) patients | miR-223 mimics the downregulation of TLR4 expression in primary macrophages while downregulating the expression of FBXW7, a well-described suppressor of Toll-like receptor 4 (TLR4) signaling. | [97] |
| miR-1, -133a, -26b, -208b, -499, | In vivo | Peripheral blood mononuclear cells from 152 hypertensive patients and 30 healthy volunteers | Hypertensive patients showed significantly lower miR-133a expression levels (p < 0.001) and higher expression levels of miR-26b (p = 0.037), miR-1 (p = 0.019), miR-208b (p = 0.016), miR-499 (p = 0.033) and miR-21 (p = 0.002) compared to healthy controls. | [98] |
| miR-17-5p, -106a-5p, -106b-3p, -15a-5p, -15b-5p, -16-5p | In vivo | Case and control pairs (N = 15 pairs) selected from individuals with hypertension treatment | MiRs from the miR-17 and miR-15 families were downregulated in progressive chronic kidney disease with high blood pressure under hypertension treatment compared with appropriate controls. | [99] |
| miR-361-5p | In vivo | Whole blood samples from 50 paired hypertensive patients | Significant differences in hsa-miR-361-5p and hsa-miR-362-5p expression levels between samples from patients with salt-sensitive and salt-resistant hypertension (p = 0.023 and 0.049, respectively) | [100] |
| miR-34b | In vitro and in vivo | Vascular smooth muscle cells from the medial layer of the thoracic aorta collected from a total of 36 female spontaneously hypertensive and Wistar-Kyoto rats | The negative regulatory association between miR-34b and its target, CDK6, was confirmed, which has potential as a new therapeutic target in the treatment of hypertension. | [101] |
| miR-181a-5p, -27, -125a-5p, -27a-3p, -21-5p, -30a-5p, -98, -92, -22-3p, -100-5p, -99b-5p | In vitro | Human dermal microvascular endothelial cells | Significant suppression of ADM-encoded mRNA expression by endogenous miR-181a-5p, ATP2B1 by miR-27 family, FURIN by miR-125a-5p, FGF5 by let-7 family, GOSR2 by miR-27a-3p, JAG1 for miR-21-5p, SH2B3 for miR-30a-5p, miR-98, miR-181a-5p, and miR-125 family, TBX3 for miR-92 family, ADRA1B for miR-22-3p, ADRA2A by miR-30a-5p and miR-30e-5p, ADRA2B by miR-30e-5p, ADRB1 by the let-7 and miR-98 family, EDNRB by the miR-92 family, and NOX4 by the miR-92 family, miR -100-5p and miR-99b-5p (n = 3–9; p < 0.05 versus scrambled anti-miR). | [102] |
| MicroRNAs | Model | Sample Size | Main Finding | Reference |
|---|---|---|---|---|
| let-7a, -125b, -100, -21 | In vivo | Gingival tissue samples collected from 100 individuals with healthy gingiva and 100 chronic periodontitis patients | Expression analysis revealed that let-7a and miR-21 were upregulated, whereas miR-100, miR-125b, and LIN-28 were downregulated in chronic periodontitis patients relative to healthy individuals. They found that NF-κB was a common target among all four miRNAs. | [19] |
| let-7a, let-7c, -130a, miR301a, miR-520d and miR-548a, miR-181b, miR-19b, miR-23a, miR-30a, miR-let7a, miR-301a | In vivo | Normal healthy gingiva and diseased gingival tissues obtained from patients undergoing periodontal treatment | miR-let-7a, let-7c, miR-130a, miR301a, miR-520d, and miR-548a were more than 8-fold up-regulated compared to healthy gingiva. MiR-181b, miR-19b, miR-23a, miR-30a, miR-let7a, and miR-301a were successfully amplified and increased significantly more in periodontitis cases than in healthy subjects. | [103] |
| miR-1274b, -let-7b-5p, -24-3p, -19b-3p, -720, -126-3p, -17-3p, -21-3p. | In vivo | Gingival tissue samples from 9 nonsmoker individuals with chronic periodontitis and 9 nonsmoker individuals with aggressive periodontitis | No differences were observed in the expression profiles of miRNAs between aggressive periodontitis and chronic periodontitis (p > 0.05). The most expressed miRNAs in both groups were hsa-miR-1274b, hsa-let-7b-5p, hsa-miR-24-3p, hsa-miR-19b-3p, hsa-miR-720, hsa-miR-126-3p, hsa-miR-17-3p, and hsa-miR-21-3p. | [104] |
| miR-146a, -155 | In vivo | Gingival crevicular fluid from 24 healthy individuals with chronic periodontitis, 24 patients with chronic periodontitis in association with DM type 2, 24 healthy individuals with clinically healthy periodontium or 24 patients with clinically healthy periodontium in association with DM type 2 |
They revealed that miR-146a and miR-155 levels were significantly associated with periodontitis. | [105] |
| miR-17 | In vivo | Healthy human tooth samples collected from 8 individuals and teeth affected by periodontal disease collected from seven periodontics clinic patients diagnosed with chronic periodontitis | They found that inflammation resulted in an inhibition of miR-17 levels, which partly reversed the differentiation potential of mesenchymal stem cells (MSCs) isolated from periodontitis-affected periodontal ligament tissue (PDLSC). They confirmed that Smurf1 is a direct target of miR-17 in PDLSC. | [106] |
| miR-200b | In vivo | Gingival excess tissue sample was collected from obese and normal weight subjects | The miRNA profile of gingival tissue from obese patients with periodontitis, compared to normal weight patients, showed 13 upregulated and 22 downregulated miRNAs, among which miR-200b was validated by qRT-PCR for significantly increase in obesity. | [107] |
| miR-21-5p, -498, -548a-5p, -495-3p, -539-5p, -34c-3p, -7a-2-3p | In vitro | LPS-treated cells from primary human periodontal ligament isolated from explanted healthy periodontal ligament | It was identified 22 upregulated miRNAs and 28 downregulated miRNAs in the LPS-treated periodontal ligament. Seven upregulated (miR-21-5p, 498, 548a-5p) and downregulated (miR-495-3p, 539-5p, 34c-3p, and 7a-2-3p) miRNAs. | [108] |
| miR-302a-3p | In vitro | LPS-treated cells from human mandibular osteoblast-like cells and LPS-treated cells of immortalized normal oral keratinocyte | miR-302a-3p regulates RANKL expression in HMOB within the PGE2 -IFNγ regulatory network. | [109] |
| miR-543 | In vitro | Human periodontal ligament-derived stem cells | miR-543 was upregulated during osteogenic differentiation of human periodontal ligament-derived stem cells. Functional experiments showed that overexpression of miR-543 could enhance osteogenesis, while inhibiting miR-543 resulted in reduced formation of mineralized nodules. The ERBB2 transducer, 2 (TOB2) was identified as a target gene of miR-543. | [110] |
| miR-664a-3p, -501-5p, -21-3p | In vivo | Serum samples from 30 healthy patients without periodontitis and 30 patients with chronic periodontitis | The expression of hsa-miR-664a-3p, hsa-miR-501-5p, and has-miR-21-3p was higher in the periodontitis group than in the control group (p < 0.05). | [111] |
| miR-126*, -20a, -142-3p, -19a, -let-7f, -203, -17, -223, -146b, 146a, -155, -205 | In vivo | Gingival tissues obtained from 10 periodontitis patients and 10 healthy subjectshasHsa-miR-126*, hsa-miR-20a, hsa-miR-142-3p, hsa-miR-19a, hsa-lehasf, hsa-has-203, has-miR-17hassa-miR-223, hsa-miR-14has hsa-miR-146a, hsa-miR-155, and hsa-miR-205 showed differential expression levels in subjects with periodontitis in relation to healthy subjects. | [112] | |
| miR-128, -34a, -381, -15b, -211, -372, -656 | In vivo and in vitro | Gingival tissues from periodontitis patients and healthy subjects THP-1 cells and CA9-22 challenged with Porphyromonas gingivalis |
The gingival tissues of patients with periodontitis showed a higher expression of miRNA-128, miRNA-34a, and miRNA-381 and a decrease in the expression of miRNA-15b, miRNA-211, miRNA-372, and miRNA-656. | [113] |
| miR-150, -223, -200b, -379, -199a-5p, -214. | In vitro | Primary human gingival fibroblasts obtained from patient gingival connective tissue explants | The most overexpressed miRNAs (by >2.72 times) were hsa-miR-150, hsa-miR-223 and hsa-miR-200b, and the three most underexpressed miRNAs (by <0.39 times) were hsa-miR-379, hsa-miR-199a-5p and hsa-miR-214 in inflamed gums of patients with periodontitis. | [114] |
| miR-451, -223, -486-5p, -3917, -1246, -1260, -141, -1260b, -203, -210, -205 | - | Data obtained from Gene Expression Omnibus database | Four miRNAs (hsa-miR-451, hsa-miR-223, hsa-miR-486-5p, hsa-miR-3917) were significantly overexpressed and 7 (hsa-miR-1246, hsa-miR-1260, hsa-miR-141, hsa-miR-1260b, hsa-miR-203, hsa-miR-210, hsa-miR-205 *) were underexpressed by >2 times in diseased gums of patients with periodontitis versus healthy gums. | [115] |
| miR-200c | In vivo and in vitro | 12-week-old male Sprague Dawley rats microinjected with LPS-PG into the gingival sulcus Primary human gingival fibroblasts |
They confirmed that local treatment with miR-200c effectively protected alveolar bone resorption in a rat model of periodontitis, by reducing the distance between the cementum-enamel junction and the alveolar bone crest and the interroot space in the second upper molar. | [116] |
This entry is adapted from the peer-reviewed paper 10.3390/ijms25041992
