As an important barrier between the cytoplasm and the microenvironment of the cell, the cell membrane is essential for the maintenance of normal cellular physiological activities. An abnormal cell membrane is a crucial symbol of body dysfunction and the occurrence of variant diseases; therefore, the visualization and monitoring of biomolecules associated with cell membranes and disease markers are of utmost importance in revealing the biological functions of cell membranes. Due to their biocompatibility, programmability, and modifiability, DNA nanomaterials have become increasingly popular in cell fluorescence imaging in recent years. In addition, DNA nanomaterials can be combined with the cell membrane in a specific manner to enable the real-time imaging of signal molecules on the cell membrane, allowing for the real-time monitoring of disease occurrence and progression.
- DNA nanomaterials
- cell membrane
- fluorescence imaging
1. Introduction
2. Application of Cell Membrane Imaging
2.1. Monitoring Imaging Triggered by the Tumor Cell Microenvironment
2.1.1. ATP
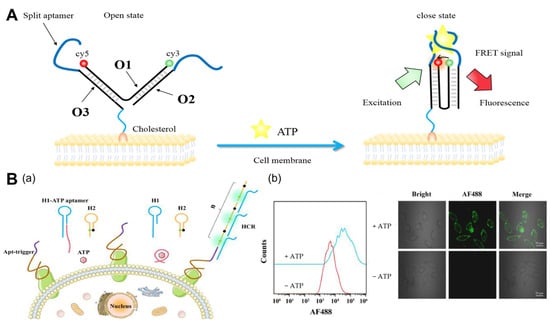
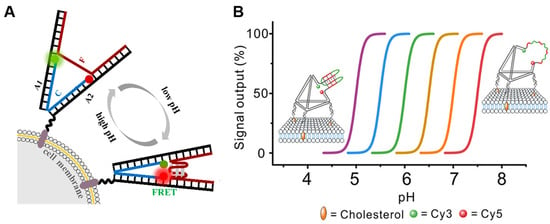
2.1.2. pH
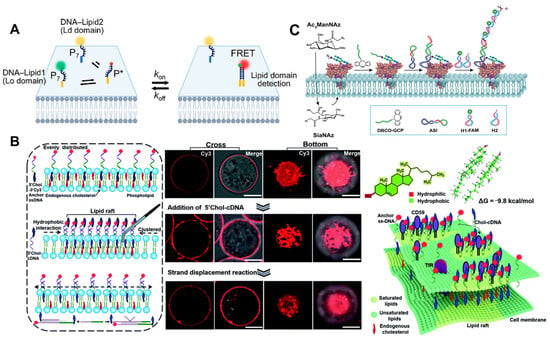
2.1.3. Metal Ions
2.2. Imaging of Cell Membrane Receptors
2.3. Imaging of Other Molecules
3. Conclusions
Although DNA nanomaterials are widely employed in cell surface fluorescence, the following obstacles must still be addressed. (1) Nowadays, most current imaging strategies involve imaging and analyzing only one or two targets, but using signal amplification techniques, such as HCR and CHA, to achieve the in situ accurate and highly sensitive imaging of multiple targets in the tumor microenvironment remains a challenge. (2) Aptamers must be designed specifically for each receptor. Currently, SELEX technology is used to screen all aptamers, and there are still receptors without aptamers. (3) The majority of DNA nanomaterials are attached to the cell membrane via covalent or noncovalent methods [44,85] but are susceptible to endocytosis. Even though some scientists have developed polymer molecular skeletons to increase the anchoring time of probes in cell membranes, long-term stability remains a problem. Monitoring intra- and extracellular signaling, cellular morphology, and structural changes requires prolonged in situ imaging, especially for slow-response events such as apoptosis. The creation of probes that can remain attached to the cell membrane for extended durations without being endocytosed remains an area of active research. (4) DNA nanomaterials are used not only for the fluorescence imaging of cell membranes but also for the functional regulation of cell membrane receptors. How to realize the integration of long-term imaging and the functional regulation of cell membranes should also be investigated. (5) FRET is dependent not only on the donor (D)–acceptor (A) separation distance and energetic resonance (i.e., D–A spectral overlap), but also on the orientation of the D emission and A absorption transition dipole moments [86,87].
This entry is adapted from the peer-reviewed paper 10.3390/molecules29010267
