Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Green & Sustainable Science & Technology
Photo-fermentation is an efficient hydrogen production pathway in which purple non-sulfur bacteria (PNSB) play an active role and produce hydrogen as a part of their metabolism under optimal conditions. These bacteria work under the influence of light to advance their metabolism and use various substrates, such as simple sugars and volatile fatty acids, to produce hydrogen.
- anaerobic conditions
- hydrogen production
- organic substrates
- volatile fatty acids
1. Photo-Fermentative Bacteria for Hydrogen Production
The production of hydrogen by photo-fermentation was recognised in the year 1949 by Gest and Kamen as it produces hydrogen with high purity without generating oxygen [14]. In this process, the organic matter is decomposed by photosynthetic bacteria in the presence of light to produce hydrogen through a reaction catalyzed by nitrogenase.
C6H12O6 + 6 H2O → 6 CO2 + 12 H2 (photosynthetic bacteria)
Bacteria commonly used for photo-fermentation include purple non-sulfur bacteria (PNSB) because they have high metabolic flexibility and can grow as photoautotrophs and chemoheterotrophs. They do not require water, unlike plants, algae, and cyanobacteria, nor do they require much free energy for hydrogen production, so no oxygen is produced in the system. Inorganic ions, carbon compounds or hydrogen are the electron sources for carrying out metabolic activities, and the generation of energy is produced by photosynthesis. PNSB show a versatile ability to utilize a wide range of organic carbon compounds for metabolic activity including pyruvate, acetate, amino acids, alcohols, organic acids, and carbohydrates. In particular, certain species within this group can use C1 compounds, such as formate and methanol, as viable carbon sources. In addition, PNSB can also utilize aromatic organic compounds, such as benzoate, cinnamate, and chlorobenzoate, to meet their carbon requirements [15]. PNSB also demonstrate the ability to assimilate a range of organic acids, including acetic, butyric, propionic, malic, and lactic acids, which becomes relevant when using organic waste as a substrate for hydrogen production. During the acidogenic phase of anaerobic digestion, PNSB can use these organic acids as carbon sources for their conversion to hydrogen and carbon dioxide. Thus, the main significance and advantage of photo-fermentative bacteria is their ability to achieve high hydrogen yields. The theoretical yield achieved from 1 mol of glucose is 12 mol H2, as given in equation 1 above, but the yield can vary slightly depending on the type of microbe. As shown in Figure 2, the following PNSB species show the ability to produce hydrogen using photo-fermentation: Rhodobacter species [16], such as Rhodobacter sphaeroides [17] and Rhodobacter capsulatus [18]; Rhodopseudomonas species [19], such as Rhodopseudomonas palustris [20], Rhodopseudomonas capsulata; Rhodospirillum rubrum [21]; Rhodovulum species, including Rhodovulum sulfidophilum [22]. These microbes follow the pathways supported by ATP-dependent nitrogenase, as the metabolic pathway is similar in all these bacteria; among them, Rhodospirillum rubrum was reported as the first PNSB to produce hydrogen.
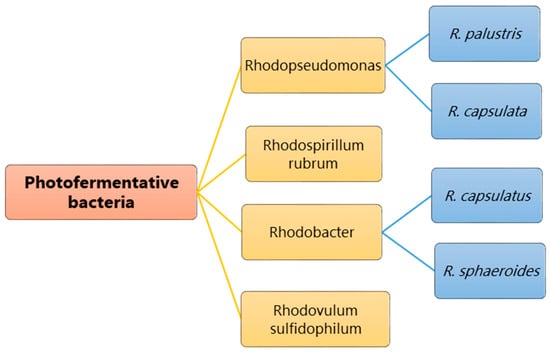
Figure 2. Bacteria mostly studied for photo-fermentative hydrogen production.
2. Metabolic Pathway of Photo-Fermentative Bacteria
Hydrogen metabolism is of great importance in maintaining the stability and performance of various microbial biotopes at the ecosystem level. Hydrogen has been used as an electron donor for reductive dehalogenation by several microbes and for the presence of hydrogenase enzymes [23]. There are certain key factors associated with the metabolic pathways of these microbes that enhance hydrogen production. These include light absorption, as these bacteria possess photosynthetic pigments that allow light energy to be absorbed from the environment, followed by the incorporation of organic substrates, such as sugars and volatile fatty acids, which are then catabolized through metabolic pathways.
This light energy enables the reduction of protons to produce molecular hydrogen while carbon fixation takes place for bacterial growth. R. palustris, which has been extensively studied for hydrogen production, assimilates carbon dioxide into organic compounds via the Calvin–Benson–Bassham cycle, which contributes to bacterial growth and its biomass production, although this cycle is not the primary route of hydrogen production in this species [24]. The photosystem of PNSB is not very efficient at splitting water, so no oxygen is involved, making it suitable for hydrogen production. Protons are pumped into the periplasmic space, as electrons move through the electron transport chain to ferredoxin, which delivers these electrons to the nitrogenase enzyme to reduce molecular nitrogen to ammonia, creating a proton gradient or proton motive force. This is followed by the hydrogenase enzyme present in these bacteria, which catalyzes the reversible reaction of proton reduction to form molecular hydrogen, which accumulates in the periplasmic space. In the absence of nitrogen, nitrogenase functions similarly to hydrogenase, catalyzing the reduction of protons with the electrons derived from ferredoxin.
The processes of hydrogen production and efflux generate ATP as a form of energy through a process called chemiosmotic ATP synthesis, which is used by the cells for various metabolic processes [25]. A general outline of the metabolic pathway followed by these photo-fermentative bacteria is presented in Figure 3, and a detailed description of the complete process is given in Figure 4.
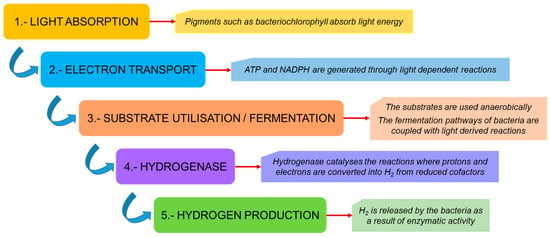
Figure 3. A general outline of hydrogen production by photo-fermentative bacteria.
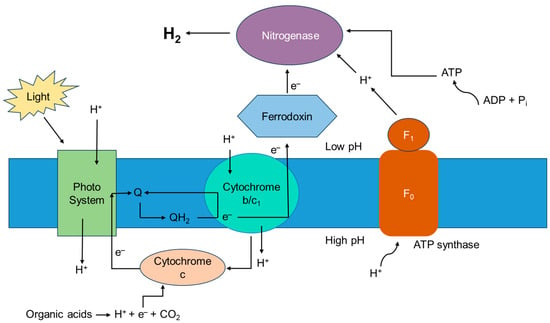
Figure 4. Photo-fermentative hydrogen production in PNSBs. Adapted with permission from Ref. [26]. 2012, Androga, Özgür, Eroglu, Gündüz, Yücel—IntechOpen Limited.
3. Rhodopseudomonas
Rhodopseudomonas bacteria contain a reaction center with bacteriochlorophyll b [27], which was discovered in 1963 and has a wide range of metabolic processes. It is found in a variety of marine environments as well as in soil [28]. Some strains of these microbes are used in photo-fermentation and will be discussed here.
3.1. Rhodopseudomonas palustris
This is a Gram-negative bacterium that uses light energy for its metabolic pathway, which leads to the conversion of organic matter into hydrogen. These microbes can alleviate heavy metal toxicity, improve plant growth, and control various plant pathogens [28].
Several studies have been carried out using these bacteria to analyze hydrogen production during photo-fermentation. In a study conducted by Wang et al., the biochemical kinetics of photo-fermentative hydrogen production were investigated experimentally and numerically in order to optimize the photo-fermentative hydrogen production process. R. palustris showed a maximum specific growth rate of 0.26 h−1 at an irradiance of 47.4 W/m2, 30 °C culture temperature, at 12 h and a pH of 7.0. An initial glucose concentration of 9.9 g/L was used as a substrate in a batch mode study, and gas chromatography was performed to analyze the hydrogen content with the flow rate of argon as carrier gas adjusted to 25 mL/min; a maximum specific hydrogen production rate of 1.63 × 10−3 h−1 was observed. An increase in the specific hydrogen production rate was obtained with an increment in temperature to 30 °C, but this decreased to 1.11 × 10−3 h−1 when the temperature was further increased to 40 °C, suggesting a reversible inactivation of cellular enzymes and inhibition of hydrogen formation [29].
In a further analysis, mutant strains of R. palustris were synthesized by the manipulation of nitrogen regulatory genes, as it proved to be an effective way of increasing the ammonia tolerance in photosynthetic bacteria to improve hydrogen production. Acetate (20 mM) and butyrate (20 mM) were used as carbon sources for cell growth and hydrogen production at 30 °C in Sistrom’s medium with an irradiance of 39.5 W/m2. The mutant strain, named nifA draT2, was 25 times more tolerant to ammonium than the wild-type strain, demonstrating that manipulation of nitrogen regulatory genes is a reliable way to increase the ammonium tolerance of these bacteria and improve their hydrogen-producing capacity. The hydrogen produced was collected in syringes and analyzed by gas chromatography. The mutant strain nifA draT2 yielded 2744 ± 66 mL of hydrogen/L, which was higher than the wild-type strain, indicating that the hydrogen production was enhanced by the nifA mutation [30].
The review by Sagir and Alipour discussed the immobilization of photosynthetic bacteria, which helped to increase hydrogen production, and different materials, such as agar, glass beads, alginate, and porous glass have been used for the attachment and immobilization of R. palustris in different studies [31]. A strategy to increase the productivity of hydrogen was recently analyzed in R. palustris using lignocellulose substrate by supplementing it with iron and molybdenum, which acts as a biocatalyst to facilitate electron transfer and ultimately an increase in hydrogen production with a maximum of 2.15 mM/h [32].
The results of several studies showed that the optimum process conditions for maximizing hydrogen yields from R. palustris strains were close to pH 7 and at a temperature range of 30–40 °C in different bioreactors and varying light intensities from different sources. Therefore, this microbe has the potential to use different organic sources to produce hydrogen, offering a glimpse into the near future where sustainable and renewable energy sources will play an important role.
3.2. Rhodopseudomonas capsulata
R. capsulata can produce hydrogen by converting light into chemical energy. This microbe is mainly found in aquatic environments and is known for its ability to perform anoxygenic photosynthesis, as it does not produce oxygen as a by-product. Bacteriochlorophyll and carotenoids are the essential pigments for photosynthesis in these bacteria.
In a study conducted by Eroglu et al., the maximum hydrogen productivity was achieved at a temperature of 30 °C and an irradiance of 31.6 W/m2 [49]. In addition, a study carried out in 1971 to elucidate the photophosphorylation system of this microbe found that sonication of chromatophores in the presence of EDTA produced non-photo-phosphorylating particles and protein factors that helped to restore photophosphorylation [50].
It was observed that the metabolism of R. capsulata was affected by intermittent illumination, suggesting that the light conversion and biosynthesis are closely linked in these bacteria. Thus, the analysis of R. capsulata could help to gain an insight into the phototrophy of the bacteria and its ability to produce hydrogen, and to identify the relationship between the energy conversion and biosynthesis. R. capsulata has been identified for its ability to degrade toxic contaminants along with hydrogen. Benzoate is one of the identified inhibitory compounds that can prevent hydrogen production during anaerobic metabolism in the absence of light and electron acceptors, but under phototrophic conditions, R. capsulata can extract hydrogen and use benzoate as a carbon source [51].
The studies showed that a mixture of acetate, propionate, and butyrate is an optimal carbon source for R. capsulata strains to produce hydrogen in the PBRs. However, the microbe has not been analyzed in many studies but has shown a great potential for photo-biological hydrogen production, paving the way for the production of renewable energy sources in the future.
3. Rhodobacter
Rhodobacter is a Gram-negative bacterium belonging to the family Rhodobacteraceae, which has a 16S rRNA gene-based phylogeny and is known for its anoxygenic photosynthetic ability [52]. They are found in freshwater and marine habitats. This genus comprises a heterogeneous group of members, and the analysis of its pan-genome revealed that 1239 core genes are shared by 12 Rhodobacter [55].
3.1. Rhodobacter capsulatus
R. capsulatus is a member of the PNSB. Like other microbes in the genus Rhodobacter, these bacteria can also grow and function by using energy from light without releasing oxygen as a by-product [56]. These bacteria are facultative anaerobes that can switch metabolic pathways accordingly and can use a variety of organic compounds as carbon and energy sources. In these bacteria, bacteriochlorophyll absorb light energy to initiate photosynthesis, electrons are transferred from water to the photosynthetic reaction centers within a cell, and in return, protons are pumped into the periplasmic space, thus creating a proton gradient. The hydrogenase enzyme catalyzes the proton reduction reaction to produce hydrogen gas as the end product [57].
Ma et al. reduced the bacteriochlorophyll content of R. capsulatus and used genetic engineering and transposon mutagenesis to create a mutant strain that produces 50.5% more hydrogen. The reduction in pigment allows more light to reach the bacteria, resulting in increased phototrophic growth and hydrogen production [58].
3.2. Rhodobacter sphaeroides
R. sphaeroides is a photosynthetic bacterium found in freshwater and marine ecosystems with a metabolism that produces hydrogen under illumination in the presence of an inert, anaerobic atmosphere. The conditions for hydrogen production must be carefully adjusted, but sometimes it is unavoidable and an alternative mode prevails, and the microbe can modify its metabolic pathways due to its versatile metabolism [68].
These bacteria, like other PNSBs, have bacteriochlorophyll, which absorbs photons and converts them into chemical energy. NiFe hydrogenases, which catalyze the formation of molecular hydrogen, are commonly found in these bacteria.
R. sphaeroides has been analyzed in several studies with different carbon sources and has shown great potential as a renewable and environmentally friendly source with the ability to convert organic compounds into hydrogen gas. Further optimization of the environmental conditions for its growth and genetic engineering of its strains helps to enhance its hydrogen production capabilities, making this microbe an efficient candidate for the development of green energy.
4. Rhodospirillum rubrum
R. rubrum is a spiral, purple photosynthetic bacterium of the genus Rhodospirillum in the class Alphaproteobacteria. The bacteria are anoxygenic phototrophs that produce extracellular elemental sulfur instead of oxygen while harvesting light with their single complex LH1 rubrum and are found in stagnant freshwater ecosystems [79]. A carotenoid spirilloxanthin is bound to the LH1 complex, which has 16 subunits observed by electron microscopy [78]. These bacteria use similar carbon sources to other purple photosynthetic bacteria, including acetate, malate, glucose, fructose, and sucrose, while producing hydrogen [79].
Different strains of these bacteria were used in the PBRs to produce hydrogen and to determine exergy, which is the useful work potential or the available energy. In a study using R. rubrum (ATCC 10801), acetate was used as the carbon source in a 2 L Biostat (Sartorius, Goettingen, Germany) PBR at 30 °C, 1 atm, with 0–0.85 mL/min flow rates of liquid media, stirring at 150–500 rpm with a total of 540 h of continuous hydrogen production. Tungsten lamps with an average irradiance of 11.85 W/m2 were used as the light source, and gas samples were analyzed by gas chromatography. The process exergy efficiency was in the range of 14.71–22.90% under optimal conditions and 14.71–22.84% using conventional and eco-exergy concepts [80]. Another study demonstrating the efficiency of R. rubrum in the batch hydrogen production process was analyzed using different carbon sources, including formate, acetate, malate, glucose, fructose, and sucrose, to support microbial growth. The process used both conventional exergy and eco-exergy concepts to evaluate the exergy efficiency, simultaneously produced molecular hydrogen and identified acetate as the optimal substrate for hydrogen production. A pH of 7.5, 60 W tungsten lamps with a uniform irradiance of 7.9 W/m2, a pressure of 1 atm, and a constant speed of 200 rpm were used to produce hydrogen. Using acetate as a carbon source, a minimum of 189.67 and 181.40 kJ/kJ of H2 exergy destruction was obtained, proving that the exergy analysis can be used to determine and compare the renewability of hydrogen production [21].
5. Rhodovulum sulfidophilum
R. sulfidophilum is a marine acid-tolerant photo-fermentative bacterium. They are Gram-negative, rod-shaped bacteria and can accumulate poly(3-hydroxybutyrate) (PHB) to over 50% of cell dry weight when acetate is used as a substrate. Hydrogen production in the presence of PHB by R. sulfidophilum shows that the loss of reducing equivalents to produce hydrogen can be recovered by the degradation of PHB [81].
In most cases, a neutral initial pH is preferred by the photosynthetic bacteria for optimal hydrogen production, but screening for acid-tolerant strains is important to increase the production of hydrogen in an acidic environment and at higher temperatures. Thus, a Tn7-based transposon was inserted into the genomic DNA of R. sulfidophilum P5 and the hydrogen yield and average hydrogen production rate of 2.16 ± 0.10 mol/mol acetate and 10.06 ± 0.47 mL/L h were observed, respectively, which were approximately 17.32- and 15.37-fold higher than those of the wild-type strain, respectively [82]. Another strain, R. sulfidophilum TH-102, was studied, and it was analyzed individually and in co-culture with other microbes of dark fermentation; the results showed that the addition of strain TH-102 can stabilize pH, decrease oxygen reduction potential, and prolong hydrogen production. The analyses provided data showing that hydrogen production during dark and photo-fermentation alone was sustained for 72 and 168 h, but when using a co-culture it went from 168 to 216 h. The co-culture with the ratio of dark/photo-fermentation bacteria 1:10 produced the highest hydrogen yield of 1694 ± 21 mL/L [83].
Based on the studies with different photo-fermentative microbes, single-stage and two-stage fermentation have been carried out to produce hydrogen. Single-stage photo-fermentation is more cost-effective than two-stage because it utilizes a wide range of substrates, thus yielding more hydrogen as compared to dark fermentation [84]. Two-stage photo-fermentation involves either coupling dark fermentation with photo-fermentation in sequence or combining bacteria used for dark and photo-fermentation to increase hydrogen yield [85]. By dividing the hydrogen production process into two stages, each stage could be optimized based on specific requirements and could lead to an improved hydrogen production efficiency compared to single-stage photo-fermentation.
This entry is adapted from the peer-reviewed paper 10.3390/app14031191
This entry is offline, you can click here to edit this entry!
