Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Gut luminal dysbiosis and pathobiosis result in compositional and biodiversified alterations in the microbial and host co-metabolites. The primary mechanism of bacterial evolution is horizontal gene transfer (HGT), and the acquisition of new traits can be achieved through the exchange of mobile genetic elements (MGEs). Introducing genetically engineered microbes (GEMs) might break the harmonized balance in the intestinal compartment.
- horizontal gene transfer
- genetically engineered microorganisms
- mobile genetic elements
- regulation
- autoimmune diseases
- microbiome
- dysbiome
- gut
- intestinal
1. Introduction
Many essential functions of the human body depend on the enteric symbiotic microbiota composition and biodiversity, essential components for human health. This intricate host–taxa relationship is a dynamic result of their long-term coevolution. This eubiosis harmonically maintains the host’s nutrition, metabolic passways, physiology, protective immune system and even behavior to the extent that we need them and cannot live without them. Greater phyla diversity is associated with microbiota resilience, sustained stability and greater ability to perform metabolic functions. The loss of microbiota phylogenic diversity and enhanced gut dysbiotic composition were associated with the Western lifestyle and several inflammatory, neurodegenerative, neurodevelopmental, infectious, metabolic, cancer and autoimmune diseases (ADs) that put human health at risk [1][2][3][4][5].
The primary mechanism of bacterial evolution is horizontal gene transfer (HGT), and new traits can be acquired through this mobile element exchange. Introducing GEMs might break the harmonized balance in the intestinal compartment [1][6]. The stable temperature, constant physicochemical conditions, continuous food supply, extremely high concentration of prokaryotic cells and phages, and plenty of opportunities for conjugation on the surfaces of host tissues and food particles represent one of the most favorable ecological niches for GEM-originated horizontal gene exchange of detrimental and harmful genetic sequences. Newly developed techniques of bacterial-mediated drug delivery have recently emerged using genetically engineered microbes aiming to locally deliver recombinant therapeutic proteins to the human gut. They are often called live biotherapeutic products, but they deliberately embed potential risks.
2. Numerous Harmful Mobile Genetic Elements (MGEs) Can Be Transferred to the Human Microbiome
Through their genomes, bacteria are subjected to rapid mutations and numerous rearrangements or HGT among and/or within bacterial species. Those MGEs, represented by bacteriophages, transposons, plasmids, and other pathogenic islands, represent a substantial amount of the microbial genome. Applying GEMs to the intestinal lumen can annulate the expression of beneficial genes while inducing the secretion of detrimental proteins. Alternatively, the GEMs can acquire the MGEs in the gut lumen. The following are various major and harmful clinical examples (Figure 1, Table 1).
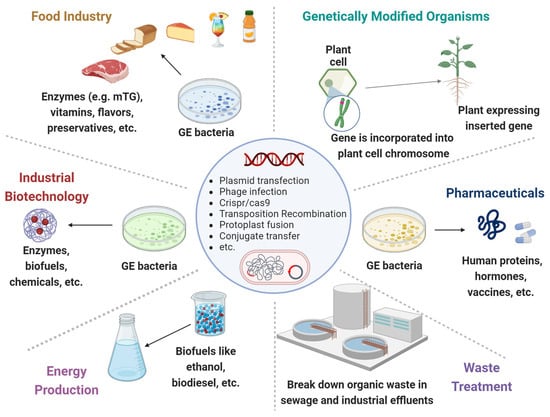
Figure 1. Genetically engineered microorganisms (GEMs) applications. GEMs have a wide range of applications across various fields due to their versatility and the precision of genetic engineering techniques. Food Industry: Production of vitamins, flavors, enzymes, and preservatives. They can help in improving the nutritional value, taste, and shelf-life of food products. Agriculture: Promote plant growth, increase nutrient uptake, and protect plants from pests and diseases. Medicine and Health Care: Cost-effective production of pharmaceuticals, including insulin, growth hormones and vaccines. Waste Treatment: Break down hazardous substances like oil spills, heavy metals and other toxic chemicals. Energy Production: The production of biofuels like ethanol and biodiesel. Industrial Biotechnology: Improve chemical production to increase yields and reduce environmental impacts.
-
Antibiotic resistance genes (ARGs) and multidrug resistance (MDR) genes are the most reported [1][7][8][9]. Less reported but not less important is the development of resistance of bacteria to phages [10][11], drug resistance to cancer therapy [12], resilience against antimicrobial defensive factors [13] and the MDR genes transfer along the food chain, including by contaminated and industrially processed nutrients [14]. The emergence of the resistome represents a worldwide health threat driven by the increasing unnecessary use of antibiotics and anticancer therapy. It occurs mainly by accumulating ARGs and MDR genes on MGEs, which is made possible by HGT [1][15]. Even the frequently consumed Lactobacillus reuteri was reported to carry ARGs [16][17]. The ARG does not originate only from human antibiotic consumption—antibiotic residue in food from animal sources can also drive the resistome [14][18]. Most recently, a high rate of ARG carried by Enterobacterales and diarrheagenic Escherichia coli in healthy donors screened for human fecal transplantation was noted [19]. The authors recommended multiplex PCR panels for stool donor screening. One wonders if the GEMs, said to benefit human health, are screened for ARGs or MDR genes.
-
Microbial-engineered enzymes are an exponentially growing area that has become indispensable to processed food production, pharmaceuticals, and numerous other commercial goods [20]. Despite their beneficial effects on the processed food industries with increased production yields and “enhancing quality and sustainability” [21], multiple scientific publications are calling for a reassessment of their safety [22][23][24][25][26]. Intriguingly, a recent call was to reevaluate the GRAS definition allocated to various processed food additive ingredients. More reliable and updated approaches are offered to enzyme and other food nutritional categories for a more scientifically rigorous, sound and transparent application of the GRAS concept [27][28][29][30][31][32]. Moreover, a call to label, declare utilization and ensure consumer transparency regarding GEM enzymes is expressed in multiple scientific publications [28][33][34][35].Many nutritional components and nutrients are treated by GEM enzymes, resulting in post-translational modified proteins, turning naïve peptides into immunogenic complexes [2][30][32][36]. There are multiple examples of genetically engineered microbial enzymes; hence, one example will be expanded, namely the microbial transglutaminase (mTG).Microbial transglutaminase is a frequently used processed food additive, and its cross-linked complexes usage is expanding exponentially. The enzyme was classified as a processing aid and was granted the GRAS (generally recognized as safe) definition decades ago, thus avoiding a thorough assessment according to current criteria of toxicity and public health safety [24][25][26][37][38][39].In contrast to the manufacturer’s declarations and claims, mTG and/or its transamidated complexes are proinflammatory, immunogenic, allergenic, pathogenic and potentially toxic, hence compromising public health [24][25][26]. Being a member of the transglutaminase family and functionally imitating the tissue transglutaminase to demidate or transamidate gliadin peptides, it was recently reported as a potential inducer of celiac disease [26][40][41][42]. In addition, its family member, the tissue transglutaminase, is a well-known inflammation inducer, fibrosis mediator and is heavily involved in sepsis [43][44]. Since mTG functionally imitates its endogenous member, one wonders if it contributes to those morbid conditions.Microbial transglutaminase and its docked complexes have numerous detrimental effects. Interestingly, in contrast to many publications showing the positive and beneficial aspects of mTG usage [45][46][47][48][49][50], there is evidence for the negative and harmful aspects of enzyme usage that might impact and compromise public health [25][26][28]. The debate between the GRAS category allocated by the FDA regulatory authorities for safe mTG consumption versus many critical scientific publications is ongoing. Several national regulatory committees have warned the public about the hazardous effects of mTGs [24][25][26]. In the case of mTG, it is possible for the gene responsible for its production to be transferred horizontally between microorganisms and even to eukaryotes [1][51][52]. Indeed, MGEs with mTG activity can potentially be transferred by HGT in between prokaryotes. Their presence in a gut luminal cellular compartment presents new opportunities for HGT, with the risk of inhabiting eukaryotic hosts [1][52][53]. One of the hypothetical scenarios is the acquisition of a classic microbial survival factor, such as a Trojan horse, against host self-defense barriers [1][25][26][54]. This gene exchange can happen through mechanisms like plasmid transfer or the incorporation of the transglutaminase gene into a MGE that can be transferred between bacteria. It is worth noting that the specific mechanisms and frequency of HGT for the mTG gene may depend on the particular microorganisms involved and the environmental conditions. The efforts to improve mTG production, thermostability and pH dependency by genetic engineering may do the opposite by enhancing the detrimental effects of the manipulated enzyme [55]. Finally, the fact that mTG is a bacterial survival factor can represent a significant positive selective pressure in the harsh, overcrowded luminal compartment [1][2], enhancing its HGT to other intestinal prokaryotic dwellers. It can be summarized that the mTG acts as a double-edged sword, protecting the microbes to survive in the gut lumen, hence compromising human health [24][25][26][54].
-
The place of probiotic consumption should be highlighted in terms of their side effects. Drug resistance remains a universal threat, and the fad of probiotic consumption, many of which contain antibiotic-resistant elements, is a major and serious health concern [56][57][58][59]. In 2023, emerging issues in probiotic safety arose. Whole-genome sequencing should be implemented to detect virulence factors, toxins, ARGs and other detrimental MGEs [60]. The clear assignment of species and strain identity risks to vulnerable populations and the need for adverse event reporting are important topics to regulate.Engineered probiotics through gene editing is an emerging domain. Despite the reported clinical benefits for inflammatory bowel disease, infectious, tumor and metabolic diseases, tight regulatory measures are lacking [61]. Engineered and naïve probiotics compete with the luminal microbiome for nutrients or ecological niches and thus might affect the diversity and composition of intestinal microbiota. Human health can be more affected by their interaction with the luminal lipid metabolism [62]. Once again, consumer transparency, visible labeling and safety regulations are far from satisfactory.
-
Genetically modified (GM) plants might possess beneficial traits like resistance to drought, pests and diseases, fighting climate change, improved agricultural and industrial production and enhanced nutrition. However, it also has a risky side to humans, animals and environmental health that should be regulated by national food security and regulatory authorities [63]. Mobile elements such as modified DNA can be laterally transferred to other recipients, spanning prokaryotes, eukaryotes and even to people [1][63]. More so, delaying tightened regulation risks facing increased GM plants, including genome-edited crops with deliberately altered and potentially harmful sequences [64][65][66]. A call for reconsideration before consumption [67], problematic and insufficient national legislation [68], risk of allergenicity [69] and consumer’s knowledge versus fears [70][71] are increasingly expressed concerning genetically modified food. Table 1 summarizes the harmful MGEs that potentially can compromise public health.
Table 1. MGEs harmful effect that can compromise public health.
| MGEs | Potential Harmful Effects | References |
|---|---|---|
| Antibiotic resistance or multidrug resistance genes |
Microbial antibiotic resistance Bacterial resistance to phages Drug resistance to cancer therapy Resilience against antimicrobial defensive factors Contaminated and industrially processed nutrients Potential entry to the human genome by HGT |
[1][2][3][4][5][6][7][8][9][10][11][12][13][14] |
| Microbial-engineered enzymic genes with MTG as an example. | Post-translational modified proteins, turning naïve peptides into immunogenic complexes Complexes are proinflammatory, allergenic, pathogenic and potentially toxic, hence compromising public health Potential inducer of celiac disease |
[2][11][24][25][26][28][33][34][36][41][42][72][73][74][75][76][77] |
| MGE presence in a gut lumen presents new opportunities for HGT, with the risk of inhabiting eukaryotic hosts | [1][52][53] | |
| Transfer of microbial survival factors against host self-defense barriers | [1][25][26][54] | |
| Improved enzyme production, thermostability and pH dependency by genetic engineering might enhance the detrimental effects of the manipulated enzyme | [55] | |
| Probiotics containing MGEs | Transfer of drug resistance Transfer of virulence factors, toxins, ARGs and other detrimental MGEs should be implemented |
[56][57][58][59][60][61] |
| Interference with the luminal lipid metabolism | [62] | |
| Genetically modified plants | Modified DNA or other MGEs can be laterally transferred to other recipients, spanning prokaryotes, eukaryotes and even people. | [1][63] |
| Genome-edited plants, like crops with deliberately altered and potentially harmful sequences, can invade the human microbiome or genome | [64][65][66][67][68][69][70][71] |
GEMs—genetically engineered microorganisms, HGT—horizontal gene transfer, MGEs—mobile genetic elements, mTG—microbial transglutaminase, ARGs—antibiotic resistance genes.
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms12020238
References
- Lerner, A.; Matthias, T.; Aminov, R. Potential effects of horizontal gene exchange in the human gut. Front. Immunol. 2017, 8, 1630.
- Lerner, A.; Aminov, R.; Matthias, T. Dysbiosis may trigger autoimmune diseases via inappropriate post-translational modification of host proteins. Front. Microbiol. 2016, 7, 84.
- Mosca, A.; Leclerc, M.; Hugot, J.P. Gut Microbiota Diversity and Human Diseases: Should We Reintroduce Key Predators in Our Ecosystem? Front. Microbiol. 2016, 7, 455.
- El Tekle, G.; Garrett, W.S. Bacteria in cancer initiation, promotion and progression. Nat. Rev. Cancer 2023, 23, 600–618.
- Shi, J. Editorial: Reviews in the impact of gut microbiota in health and disease. Front. Microbiol. 2023, 14, 1230925.
- Sitaraman, R. Prokaryotic horizontal gene transfer within the human holobiont: Ecological-evolutionary inferences, implications and possibilities. Microbiome 2018, 6, 163.
- Fredriksen, S.; de Warle, S.; van Baarlen, P.; Boekhorst, J.; Wells, J.M. Resistome expansion in disease-associated human gut microbiomes. Microbiome 2023, 11, 166.
- Crits-Christoph, A.; Hallowell, H.A.; Koutouvalis, K.; Suez, J. Good microbes, bad genes? The dissemination of antimicrobial resistance in the human microbiome. Gut Microbes 2022, 14, 2055944.
- Bag, S.; Ghosh, T.S.; Banerjee, S.; Mehta, O.; Verma, J.; Dayal, M.; Desigamani, A.; Kumar, P.; Saha, B.; Kedia, S.; et al. Molecular Insights into Antimicrobial Resistance Traits of Commensal Human Gut Microbiota. Microb. Ecol. 2019, 77, 546–557.
- Suh, G.A.; Patel, R. Clinical phage microbiology: A narrative summary. Clin. Microbiol. Infect. 2023, 29, 710–713.
- Lerner, A.; Ramesh, A.; Matthias, T. The Revival of the Battle between David and Goliath in the Enteric Viruses and Microbiota Struggle: Potential Implication for Celiac Disease. Microorganisms 2019, 7, 173.
- Emran, T.B.; Shahriar, A.; Mahmud, A.R.; Rahman, T.; Abir, M.H.; Siddiquee, M.F.R.; Ahmed, H.; Rahman, N.; Nainu, F.; Wahyudin, E.; et al. Multidrug Resistance in Cancer: Understanding Molecular Mechanisms, Immunoprevention and Therapeutic Approaches. Front. Oncol. 2022, 12, 891652.
- Yadav, M.; Pandey, R.; Chauhan, N.S. Catabolic Machinery of the Human Gut Microbes Bestow Resilience against Vanillin Antimicrobial Nature. Front. Microbiol. 2020, 11, 588545.
- Lerner, A.; Soprun, L.; Benzvi, C. Antimicrobial Resistance along the Food Chain: Contaminated and Industrially Processed Nutrients. J. Food Nutr. Health 2022, 3, 1–11.
- Ondon, B.S.; Li, S.; Zhou, Q.; Li, F. Sources of Antibiotic Resistant Bacteria (ARB) and Antibiotic Resistance Genes (ARGs) in the Soil: A Review of the Spreading Mechanism and Human Health Risks. Rev. Environ. Contam. Toxicol. 2021, 256, 121–153.
- Rosander, A.; Connolly, E.; Roos, S. Removal of antibiotic resistance gene-carrying plasmids from Lactobacillus reuteri ATCC 55730 and characterization of the resulting daughter strain, L. reuteri DSM 17938. Appl. Environ. Microbiol. 2008, 74, 6032–6040.
- Egervärn, M.; Lindmark, H.; Olsson, J.; Roos, S. Transferability of a tetracycline resistance gene from probiotic Lactobacillus reuteri to bacteria in the gastrointestinal tract of humans. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2010, 97, 189–200.
- Qamar, M.U.; Aatika; Chughtai, M.I.; Ejaz, H.; Mazhari, B.B.Z.; Maqbool, U.; Alanazi, A.; Alruwaili, Y.; Junaid, K. Antibiotic-Resistant Bacteria, Antimicrobial Resistance Genes, and Antibiotic Residue in Food from Animal Sources: One Health Food Safety Concern. Microorganisms 2023, 11, 161.
- Chuang, C.; Lee, K.C.; Wang, Y.P.; Lee, P.C.; Chang, T.E.; Huang, Y.H.; Lin, Y.T.; Hou, M.C. High carriage rate of extended-spectrum β-lactamase Enterobacterales and diarrheagenic Escherichia coli in healthy donor screening for fecal microbiota transplantation. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 1103–1113.
- Tatta, E.R.; Imchen, M.; Moopantakath, J.; Kumavath, R. Bioprospecting of microbial enzymes: Current trends in industry and healthcare. Appl. Microbiol. Biotechnol. 2022, 106, 1813–1835.
- Jangra, S.; Srivastava, S. Microbial Enzymes in Food Industries: Enhancing Quality and Sustainability. In Food Microbial Sustainability: Integration of Food Production and Food Safety; Springer Nature: Singapore, 2023; pp. 193–221.
- Pariza, M.W.; Johnson, E.A. Evaluating the safety of microbial enzyme preparations used in food processing: Update for a new century. Regul. Toxicol. Pharmacol. 2001, 33, 173–186.
- Deckers, M.; Deforce, D.; Fraiture, M.A.; Roosens, N.H.C. Genetically modified micro-organisms for industrial food enzyme production: An overview. Foods 2020, 9, 326.
- Lerner, A.; Matthias, T. Microbial Transglutaminase is Beneficial to Food Industries but a Caveat to Public Health. Med. One 2019, 4, e190001.
- Lerner, A.; Matthias, T. Processed food additive microbial transglutaminase and its cross-linked gliadin complexes are potential public health concerns in celiac disease. Int. J. Mol. Sci. 2020, 21, 1127.
- Lerner, A.; Benzvi, C. Microbial transglutaminase is a very frequently used food additive and is a potential inducer of autoimmune/neurodegenerative diseases. Toxics 2021, 9, 233.
- Faustman, C.; Aaron, D.; Negowetti, N.; Leib, E.B. Ten years post-GAO assessment, FDA remains uninformed of potentially harmful GRAS substances in foods. Crit. Rev. Food Sci. Nutr. 2021, 61, 1260–1268.
- Lerner, A.; Benzvi, C. “Let food be thy medicine”: Gluten and potential role in neurodegeneration. Cells 2021, 10, 756.
- Sewalt, V.; LaMarta, J.; Shanahan, D.; Gregg, L.; Carrillo, R. Letter to the editor regarding “GRAS from the ground up: Review of the Interim Pilot Program for GRAS notification” by Hanlon et al., 2017. Food Chem. Toxicol. 2017, 107, 520–521.
- Neltner, T.G.; Alger, H.M.; O’Reilly, J.T.; Krimsky, S.; Bero, L.A.; Maffini, M.V. Conflicts of interest in approvals of additives to food: Determined to be generally recognized as safe: Out of balance. JAMA Intern. Med. 2013, 173, 2032–2036.
- Kruger, C. The relevance of international assessments to GRAS determinations. Regul. Toxicol. Pharmacol. 2016, 79, S119–S123.
- Roberts, A.; Haighton, L.A. A hard look at FDA’s review of GRAS notices. Regul. Toxicol. Pharmacol. 2016, 79, S124–S128.
- Lerner, A.; Matthias, T. Possible association between celiac disease and bacterial transglutaminase in food processing: A hypothesis. Nutr. Rev. 2015, 73, 544–552.
- Lerner, A.; Matthias, T. Changes in intestinal tight junction permeability associated with industrial food additives explain the rising incidence of autoimmune disease. Autoimmun. Rev. 2015, 14, 479–489.
- Kaufmann, A.; Köppel, R.; Widmer, M. Determination of microbial transglutaminase in meat and meat products. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2012, 29, 1364–1373.
- Lerner, A.; Aminov, R.; Matthias, T. Transglutaminases in dysbiosis as potential environmental drivers of autoimmunity. Front. Microbiol. 2017, 8, 66.
- Hartung, T. Rebooting the generally recognized as safe (GRAS) approach for food additive safety in the US. ALTEX 2018, 35, 3–25.
- Vandenberg, L.N.; Zoeller, R.T.; Prins, G.S.; Trasande, L. Evaluating adverse effects of environmental agents in food: A brief critique of the US FDA’s criteria. Environ. Health 2023, 22, 38.
- Kolotylo, V.; Piwowarek, K.; Kieliszek, M. Microbiological transglutaminase: Biotechnological application in the food industry. Open Life Sci. 2023, 18, 20220737.
- Lerner, A.; Matthias, T. Microbial transglutaminase should be considered as an environmental inducer of celiac disease. World J. Clin. Cases 2019, 7, 3912–3914.
- Matthias, T.; Lerner, A. Microbial Transglutaminase Is Immunogenic and Potentially Pathogenic in Pediatric Celiac Disease. Front. Pediatr. 2018, 6, 389.
- Lerner, A.; Matthias, T. Microbial transglutaminase: A new potential player in celiac disease. Clin. Immunol. 2019, 199, 37–43.
- Su, T.; Qin, X.Y.; Furutani, Y. Transglutaminase 2 as a marker for inflammation and therapeutic target in sepsis. Int. J. Mol. Sci. 2021, 22, 1897.
- Elli, L.; Bergamini, C.M.; Bardella, M.T.; Schuppan, D. Transglutaminases in inflammation and fibrosis of the gastrointestinal tract and the liver. Dig. Liver Dis. 2009, 41, 541–550.
- Kieliszek, M.; Misiewicz, A. Microbial transglutaminase and its application in the food industry. A review. Folia Microbiol. 2014, 59, 241–250.
- Duarte, L.; Matte, C.R.; Bizarro, C.V.; Ayub, M.A.Z. Review transglutaminases: Part II-industrial applications in food, biotechnology, textiles and leather products. World J. Microbiol. Biotechnol. 2019, 36, 11.
- Paolella, G.; Martucciello, S.; Vasi’cvasi’c, K.; Knez, Ž.; Leitgeb, M. Transglutaminase in Foods and Biotechnology. Int. J. Mol. Sci. 2023, 24, 12402.
- Martins, I.M.; Matos, M.; Costa, R.; Lopes-da-Silva, F.; Pascoal, A.; Estevinho, L.M.; Choupina, A.B. Transglutaminases: Recent achievements and new sources. Appl. Microbiol. Biotechnol. 2014, 98, 6957–6964.
- Fuchsbauer, H.L. Approaching transglutaminase from Streptomyces bacteria over three decades. FEBS J. 2022, 289, 4680–4703.
- Miwa, N. Innovation in the food industry using microbial transglutaminase: Keys to success and future prospects. Anal. Biochem. 2020, 597, 113638.
- Ponting, C.P.; Aravind, L.; Schultz, J.; Bork, P.; Koonin, E.V. Eukaryotic signalling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J. Mol. Biol. 1999, 289, 729–745.
- Krishnan, A.; Burroughs, A.M.; Iyer, L.M.; Aravind, L. Unexpected Evolution of Lesion-Recognition Modules in Eukaryotic NER and Kinetoplast DNA Dynamics Proteins from Bacterial Mobile Elements. iScience 2018, 9, 192–208.
- Horne, T.; Orr, V.T.; Hall, J.P. How do interactions between mobile genetic elements affect horizontal gene transfer? Curr. Opin. Microbiol. 2023, 73, 102282.
- Lerner, A.; Benzvi, C.; Vojdani, A. Cross-reactivity and sequence similarity between microbial transglutaminase and human tissue antigens. Sci. Rep. 2023, 13, 17526.
- Fatima, S.W.; Khare, S.K. Effect of key regulators in augmenting transcriptional expression of Transglutaminase in Streptomyces mobaraensis. Bioresour. Technol. 2021, 340, 125627.
- Lerner, A.; Shoenfeld, Y.; Matthias, T. Probiotics: If it does not help it does not do any harm. really? Microorganisms 2019, 7, 104.
- Lerner, A.; Matthias, T. There Are Many More Cons for Probiotics. ISR Med. Assoc. J. 2020, 22, 131.
- Imperial, I.C.V.J.; Ibana, J.A. Addressing the antibiotic resistance problem with probiotics: Reducing the risk of its double-edged sword effect. Front. Microbiol. 2016, 7, 1983.
- Dou, W.; Abdalla, H.B.; Chen, X.; Sun, C.; Chen, X.; Tian, Q.; Wang, J.; Zhou, W.; Chi, W.; Zhou, X.; et al. ProbResist: A database for drug-resistant probiotic bacteria. Database 2022, 2022, baac064.
- Merenstein, D.; Pot, B.; Leyer, G.; Ouwehand, A.C.; Preidis, G.A.; Elkins, C.A.; Hill, C.; Lewis, Z.T.; Shane, A.L.; Zmora, N.; et al. Emerging issues in probiotic safety: 2023 perspectives. Gut Microbes 2023, 15, 2185034.
- Ma, J.; Lyu, Y.; Liu, X.; Jia, X.; Cui, F.; Wu, X.; Deng, S.; Yue, C. Engineered probiotics. Microb. Cell Fact. 2022, 21, 72.
- Wu, T.; Wang, G.; Xiong, Z.; Xia, Y.; Song, X.; Zhang, H.; Wu, Y.; Ai, L. Probiotics Interact with Lipids Metabolism and Affect Gut Health. Front. Nutr. 2022, 9, 917043.
- Philips, J.G.; Martin-Avila, E.; Robold, A.V. Horizontal gene transfer from genetically modified plants—Regulatory considerations. Front. Bioeng. Biotechnol. 2022, 10, 971402.
- Halford, N.G. Legislation governing genetically modified and genome-edited crops in Europe: The need for change. J. Sci. Food Agric. 2019, 99, 8–12.
- Kleter, G.A.; Peijnenburg, A.A.C.M.; Aarts, H.J.M. Health considerations regarding horizontal transfer of microbial transgenes present in genetically modified crops. J. Biomed. Biotechnol. 2005, 2005, 326–352.
- Kamle, S.; Ali, S. Genetically modified crops: Detection strategies and biosafety issues. Gene 2013, 522, 123–132.
- Grover, A.; Ashhar, N.; Patni, P. Why genetically modified food need reconsideration before consumption? J. Fam. Med. Prim. Care 2014, 3, 188–190.
- Mammadov, V.; Mustafayeva, A. The legislation of CIS countries on the issue of genetically modified products. Med. Law 2011, 30, 555–570.
- Lee, T.H.; Ho, H.K.; Leung, T.F. Genetically modified foods and allergy. Hong Kong Med. J. 2017, 23, 291–295.
- Boccia, F.; Covino, D.; Sarnacchiaro, P. Genetically modified food versus knowledge and fear: A Noumenic approach for consumer behaviour. Food Res. Int. 2018, 111, 682–688.
- Xu, R.; Wu, Y.; Luan, J. Consumer-perceived risks of genetically modified food in China. Appetite 2020, 147, 104520.
- Lerner, A.; Matthias, T. Food Industrial Microbial Transglutaminase in Celiac Disease: Treat or Trick. Int. J. Celiac Dis. 2015, 3, 1–6.
- Lerner, A.; Matthias, T. Don’t forget the exogenous microbial transglutaminases: It is immunogenic and potentially pathogenic. AIMS Biophys. 2016, 3, 546–552.
- Agardh, D.; Matthias, T.; Wusterhausen, P.; Neidhöfer, S.; Heller, A.; Lerner, A. Antibodies against neo-epitope of microbial and human transglutaminase complexes as biomarkers of childhood celiac disease. Clin. Exp. Immunol. 2020, 199, 294–302.
- Matthias, T.; Jeremias, P.; Neidhöfer, S.; Lerner, A. The industrial food additive, microbial transglutaminase, mimics tissue transglutaminase and is immunogenic in celiac disease patients. Autoimmun. Rev. 2016, 15, 1111–1119.
- Lerner, A.; Jeremias, P.; Neidhofer, S.; Matthias, T. Comparison of the Reliability of 17 Celiac Disease Associated Bio-Markers to Reflect Intestinal Damage. J. Clin. Cell. Immunol. 2017, 8, 486.
- Amirdivani, S.; Khorshidian, N.; Fidelis, M.; Granato, D.; Koushki, M.R.; Mohammadi, M.; Khoshtinat, K.; Mortazavian, A.M. Effects of transglutaminase on health properties of food products. Curr. Opin. Food Sci. 2018, 22, 74–80.
This entry is offline, you can click here to edit this entry!
