MYC amplification or overexpression is most common in Group 3 medulloblastomas and is positively associated with poor clinical outcomes. Protein arginine methyltransferase 5 (PRMT5) overexpression has been shown to be associated with tumorigenic MYC functions in cancers, particularly in brain cancers such as glioblastoma and medulloblastoma. PRMT5 regulates oncogenes, including MYC, that are often deregulated in medulloblastomas. However, the role of PRMT5-mediated post-translational modification in the stabilization of these oncoproteins remains poorly understood. The potential impact of PRMT5 inhibition on MYC makes it an attractive target in various cancers. PRMT5 inhibitors are a promising class of anti-cancer drugs demonstrating preclinical and preliminary clinical efficacies.
- brain cancer
- medulloblastoma
- MYC
- PRMT5 inhibitors
- SDMA
1. Introduction
2. PRMT5 Structure, Function, and Localization
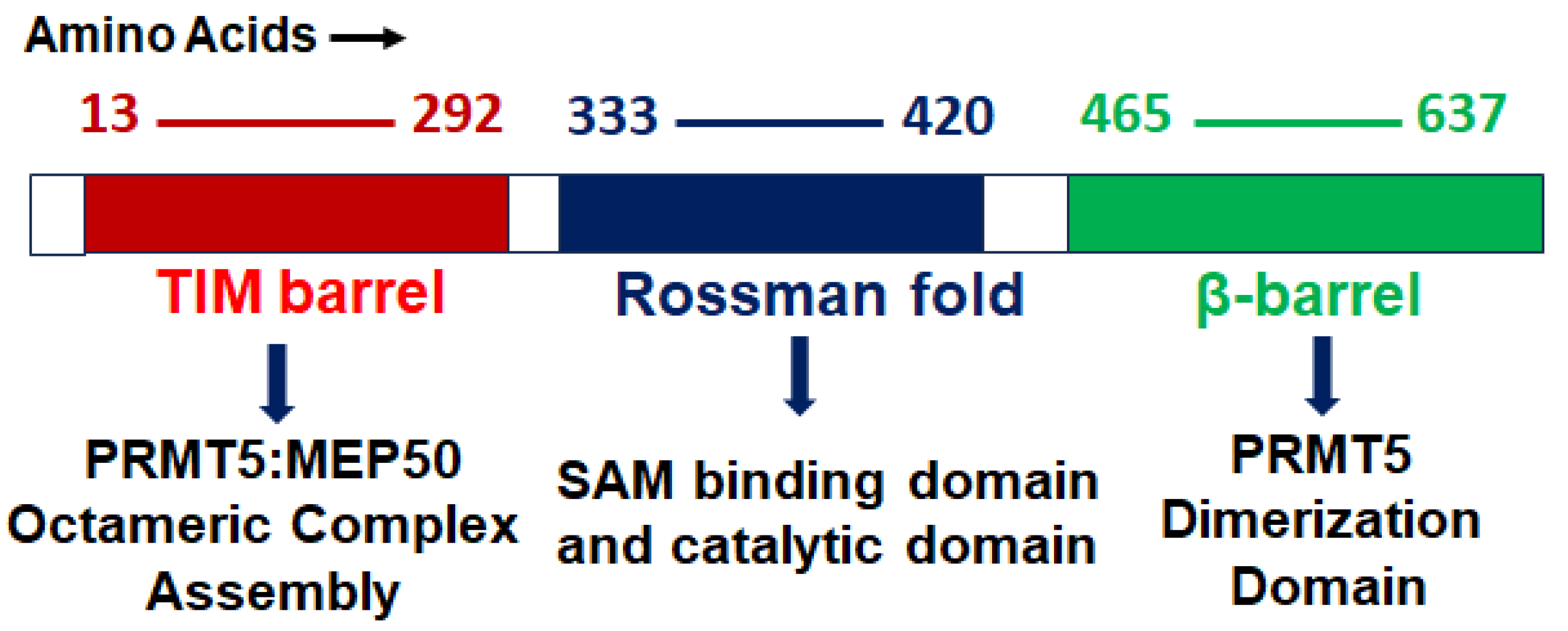
2.1. Structure
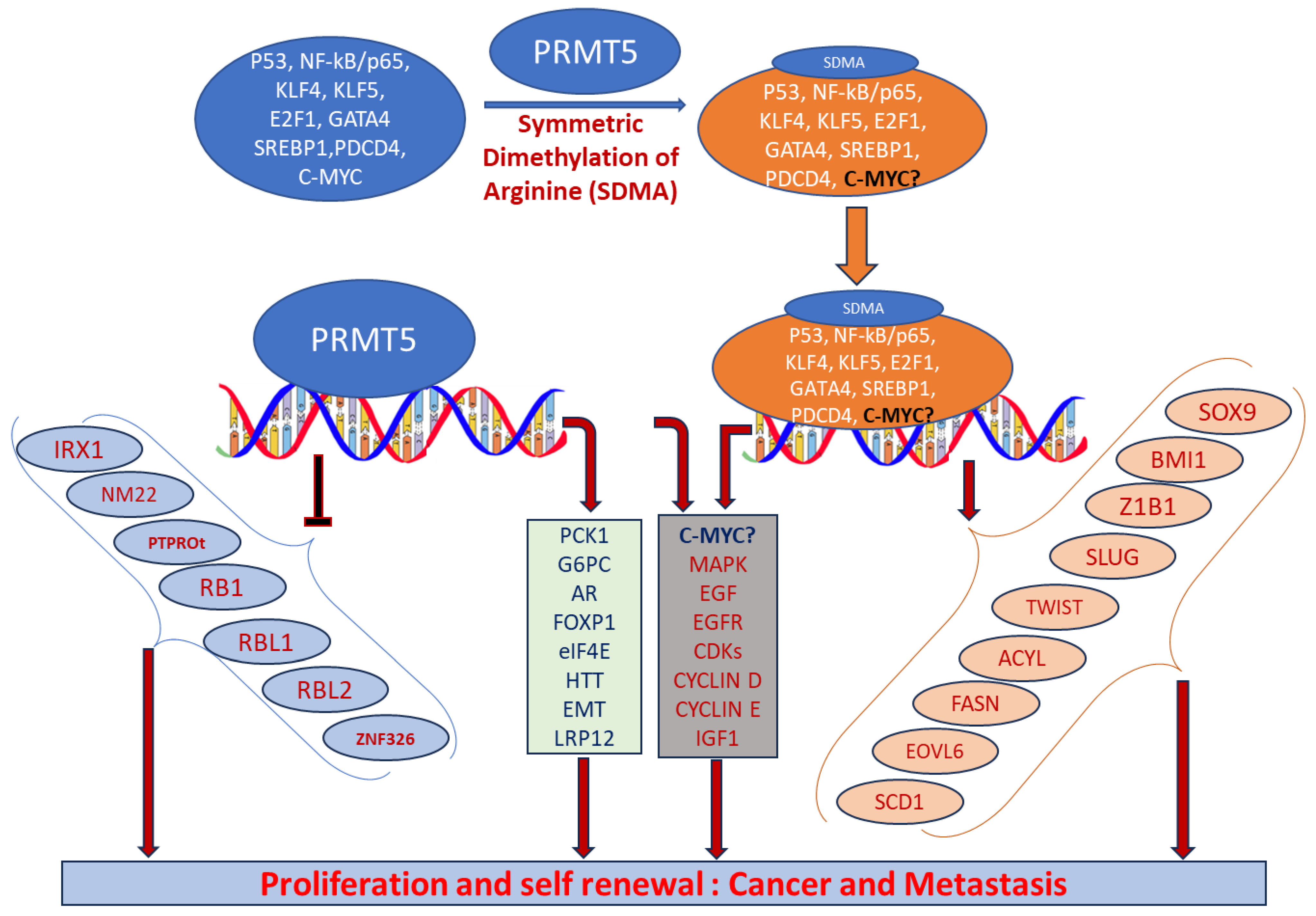
2.2. Function
2.3. Localization
|
Organ |
Cellular Function |
Mechanism |
References |
|---|---|---|---|
|
Brain |
Cell cycle progression, apoptosis |
Altered expression and stability of MYC |
[22] |
|
Phase separation |
Methylation of FUS |
[69] |
|
|
GSK3β-NF-kβ signaling |
Altered expression of E2F1 |
[70] |
|
|
HTT toxicity |
Altered expression of HTT |
[71] |
|
|
AKT-ERK signaling, cell cycle progression |
Altered expression of PTEN |
[72] |
|
|
Cell cycle progression, stemness |
Altered RNA Splicing |
||
|
mTOR signaling |
Methylation (hnRNPA1) |
[75] |
|
|
DNA instability response |
Altered expression of RNF168 |
[76] |
|
|
Cell migration, cell cycle progression, and apoptosis |
Altered expression of LRP12 |
[62] |
|
|
AKT signaling and metastasis |
Methylation of PKB |
[77] |
|
|
Lungs |
Metastasis |
Altered expression of EMT genes |
[78] |
|
Metastasis |
Altered expression of SHARPIN |
[79] |
|
|
Metastasis |
Altered expression of FGFR3/miR-99 family |
[80] |
|
|
Metastasis |
Methylation of KLF5 |
[81] |
|
|
Liver |
Lipid metabolism |
Methylation of SREBP |
[47] |
|
ERK signaling |
Altered expression of BTG2 |
[82] |
|
|
PRMY5 deprivation |
PRMT5 activity of LINC01138 |
[83] |
|
|
WNT-β-Catenin signaling |
Altered cofactor binding of LYRIC |
[84] |
|
|
Spleen |
NA |
Altered stability of MYC |
[85] |
|
Pancreas |
Glucose metabolism, cell cycle progression |
Altered stability of MYC |
|
|
Bone |
Type I interferon signaling |
Altered expression (interferon gene) |
|
|
Prostate |
AR, ERG signaling |
Altered methylation (AR) |
|
|
Ovary |
NA |
Altered methylation (E2F1) |
[92] |
|
Heart |
Transcriptional activity |
Methylation (GATA4) |
[45] |
|
Breast |
Stemness |
Altered expression of C-MYC, KLF4, and OCT4 |
[93] |
|
Stemness |
Altered expression of FOXP1 |
[94] |
|
|
Metastasis and invasion |
Altered expression of AKT genes |
[78] |
|
|
Metastasis and AKT signaling |
Methylation of AKT |
[95] |
|
|
Cell cycle progression |
Methylation of KLF4 |
[96] |
|
|
Cell migration |
Methylation of ZNF326 |
[97] |
|
|
NA |
Methylation of PDCD4 |
[98] |
Abbreviations: AKT-ERK, alpha serine/threonine-protein-extracellular-regulated kinase; AR, androgen receptor; E2F1, E2 promoter binding factor 1; EMT, epithelial–mesenchymal transition; ERG, ETS-related gene; FGFR3, fibroblast growth factor 3; FOXP1, forkhead box protein P1; FUS, fused in sarcoma; GSK3β-NF-kβ, glycogen synthase kinase; hnRNPA1, heterogeneous nuclear ribonucleoprotein A1; HTT, huntingtin protein; KLF4, Kruppel-like factor 4; LRP12, low-density lipoprotein receptor-related protein 12; LINC01138, long non-coding RNA; miR-99, microRNA99; mTOR, mammalian target of rapamycin; OCT4, octamer binding protein 4; PKB, protein kinase B; PDCD4, program cell death protein 4; SREBP, sterol regulatory element-binding protein; ZNF326, zinc finger protein 326.
3. PRMT5 Association with MYC-Driven Medulloblastoma
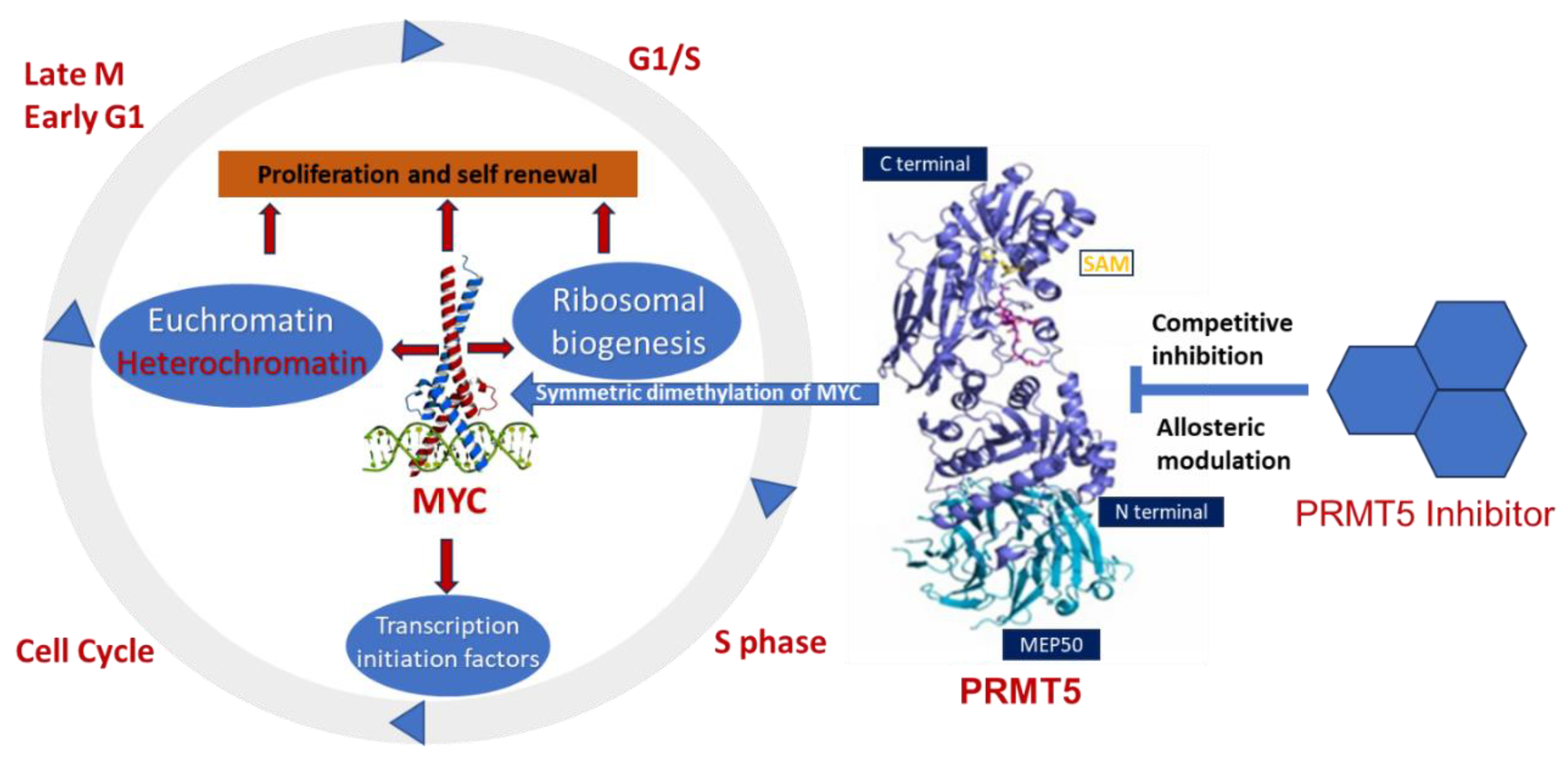
4. Potential Inhibitors of PRMT5
|
Compound Name |
Structure |
Function |
IC50 In Vitro |
In Vivo Activity |
References |
|---|---|---|---|---|---|
|
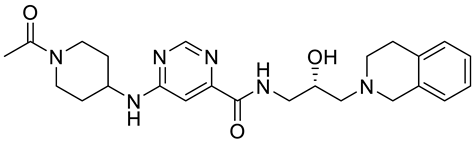
JNJ64619178 |
|
Dual SAM/substrate competitive |
0.2 nM |
Antitumor effect in lung cancer, AML, non-Hodgkin lymphoma cell line mouse xenograft |
[108] |
|
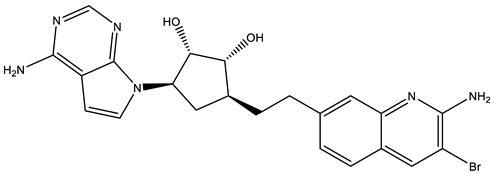
PF06939999 |
|
SAM competitive |
3.3 nM |
Antitumor effect in lung cancer |
[109] |
|
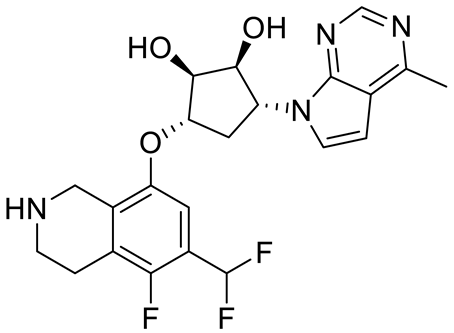
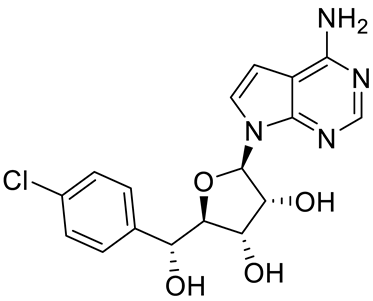
GSK3235025 EPZ015666 |
|
Substrate competitive |
22 nM |
Antitumor effect in MCL, MM, AML, GBM, and bladder cell line mouse xenografts and in a TNBC PDX mouse model |
|
|
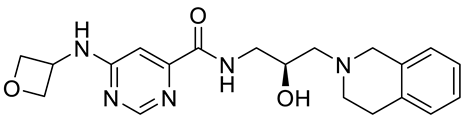
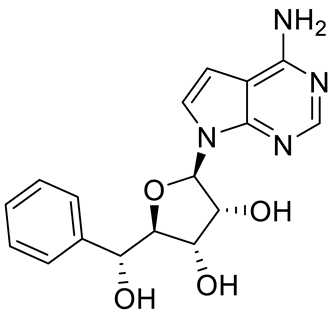
GSK591 (EPZ015866) |
|
Substrate competitive |
4 nM |
Antitumor effect in glioblastoma |
[110] |
|
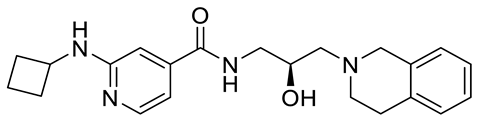
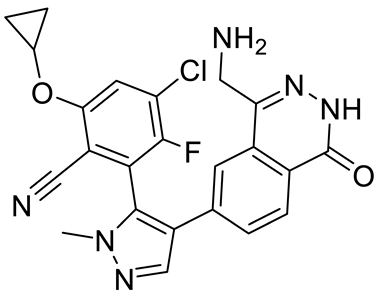
GSK3326595 |
|
Substrate competitive |
6.2 nM |
Antitumor effect in non-Hodgkin lymphoma cell line mouse xenograft and antitumor effect in a DLBCL PDX mouse model |
|
|
AMG 193 |
Structure undisclosed |
MTA cooperative inhibitor |
NA |
Antitumor effect on advanced/metastatic solid tumors |
[119] |
|
PRT543 |
|
SAM competitive |
10.8 nM |
Antitumor effect on advanced solid tumors and hematologic malignancies |
[120] |
|
PRT382 |
Structure undisclosed |
SAM competitive |
2.8 nM |
Antitumor effect on hematological tumors |
[85] |
|
PRT811 |
Structure undisclosed |
SAM competitive |
3.9 nM |
Antitumor effect on advanced solid tumor, Glioblastoma, CNS Lymphoma |
[121] |
|
TNG908 |
Structure undisclosed |
MTA cooperative inhibitor |
110 nM |
Antitumor effect on Glioblastoma, |
[122] |
|
MRTX1719 |
|
PRMT5–MTA complex inhibitor, MTA competitive |
12 nM |
Antitumor effect on solid tumor |
|
|
LLY-283 (C220) |
|
SAM competitive |
22 nM |
Reduced acute graft versus host disease incidence in mice, antitumor effect in MPN xenografts |
|
|
Compound1a |
|
Allosteric modulator |
16 nM |
Antitumor effect in breast cancer |
[127] |
|
CMP5 |
Structure undisclosed |
SAM competitive |
25 µM |
Antitumor effect in breast cancer and glioblastoma |
[72] |
|
JBI-778 |
Structure undisclosed |
Substrate competitive |
27 to 700 nM |
Antitumor effect in glioblastoma |
[128] |
|
SH3765 |
Structure undisclosed |
Substrate competitive |
NA |
Antitumor effect on advanced malignant tumors, including solid tumors and non-Hodgkin lymphoma |
[129] |
|
SCR6920 |
Structure undisclosed |
Substrate competitive |
NA |
Antitumor effect on advanced malignant tumor including solid tumor and non-Hodgkin lymphoma |
[129] |
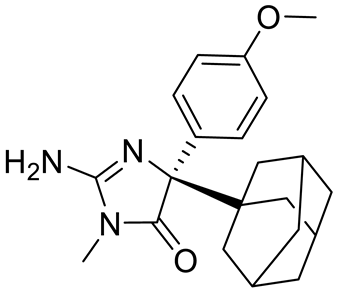
Abbreviations: AML, acute myeloid leukemia; MCL, mantle cell lymphoma; MM, myelomonocytic leukemia; GBM, glioblastoma; TNBC, triple-negative breast cancer, PDX, patient-derived xenograft; DLBCL, diffuse large B cell lymphoma; CNS, central nervous system; MPN, myeloproliferative neoplasm; nM, nano molar; NA, not available; SAM, S-adenosylmethionine; MTA, methylthioadenosine.
|
ClinicalTrials.gov Identifier |
Name of Inhibitor |
Status |
Disease |
|---|---|---|---|
|
NCT03573310 |
JNJ64619178 |
Phase I |
Neoplasm solid tumors, non-Hodgkin lymphoma, and myelodysplastic syndrome |
|
NCT03854227 |
PF06939999 |
Phase I |
Advance and metastatic solid tumors |
|
NCT03614728 |
GSK3326595 |
Phase I and II |
Metastatic solid tumors and acute myeloid leukemia |
|
NCT02783300 |
GSK3326595 |
Phase I |
Solid tumors and non-Hodgkin lymphoma |
|
NCT04676516 |
GSK3326595 |
Phase II |
Early-stage breast cancer |
|
NCT03886831 |
PRT543 |
Phase I |
Advanced solid tumors and hematological malignancies |
|
NCT05275478 |
TNG908 |
Phase I and II (recruiting) |
Locally advanced solid tumors |
|
NCT04089449 |
PRT811 |
Phase I (recruiting) |
Advanced solid tumors, recurrent glioma, and CNS lymphoma |
|
NCT05245500 |
MRTX1719 |
Phase I and II (recruiting) |
Mesothelioma, NSCLC, malignant peripheral nerve sheath tumors, solid tumors, and pancreatic adenocarcinoma |
|
NCT05094336 |
AMG 193 |
Phase I and II (recruiting) |
Advanced MTAP-null solid tumors |
|
NCT05528055 |
SCR6920 |
Phase I (recruiting) |
Advanced malignant tumors |
|
NCT05015309 |
SH3765 |
Phase I (not yet Recruiting) |
Advanced malignant tumors |
This entry is adapted from the peer-reviewed paper 10.3390/cancers15245855
References
- Packer, R.J.; Macdonald, T.; Vezina, G.; Keating, R.; Santi, M. Medulloblastoma and primitive neuroectodermal tumors. Handb. Clin. Neurol. 2012, 105, 529–548.
- Cavalli, F.M.G.; Remke, M.; Rampasek, L.; Peacock, J.; Shih, D.J.H.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morrissy, A.S.; et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017, 31, 737–754.e736.
- Northcott, P.A.; Buchhalter, I.; Morrissy, A.S.; Hovestadt, V.; Weischenfeldt, J.; Ehrenberger, T.; Grobner, S.; Segura-Wang, M.; Zichner, T.; Rudneva, V.A.; et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017, 547, 311–317.
- Taylor, M.D.; Northcott, P.A.; Korshunov, A.; Remke, M.; Cho, Y.J.; Clifford, S.C.; Eberhart, C.G.; Parsons, D.W.; Rutkowski, S.; Gajjar, A.; et al. Molecular subgroups of medulloblastoma: The current consensus. Acta Neuropathol. 2012, 123, 465–472.
- Kool, M.; Korshunov, A.; Remke, M.; Jones, D.T.; Schlanstein, M.; Northcott, P.A.; Cho, Y.J.; Koster, J.; Schouten-van Meeteren, A.; van Vuurden, D.; et al. Molecular subgroups of medulloblastoma: An international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012, 123, 473–484.
- Min, H.S.; Lee, J.Y.; Kim, S.K.; Park, S.H. Genetic grouping of medulloblastomas by representative markers in pathologic diagnosis. Transl. Oncol. 2013, 6, 265–272.
- Juraschka, K.; Taylor, M.D. Medulloblastoma in the age of molecular subgroups: A review. J. Neurosurg. Pediatr. 2019, 24, 353–363.
- Gajjar, A.J.; Robinson, G.W. Medulloblastoma-translating discoveries from the bench to the bedside. Nat. Rev. Clin. Oncol. 2014, 11, 714–722.
- Cho, Y.J.; Tsherniak, A.; Tamayo, P.; Santagata, S.; Ligon, A.; Greulich, H.; Berhoukim, R.; Amani, V.; Goumnerova, L.; Eberhart, C.G.; et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J. Clin. Oncol. 2011, 29, 1424–1430.
- Hill, R.M.; Kuijper, S.; Lindsey, J.C.; Petrie, K.; Schwalbe, E.C.; Barker, K.; Boult, J.K.; Williamson, D.; Ahmad, Z.; Hallsworth, A.; et al. Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer Cell 2015, 27, 72–84.
- Pei, Y.; Moore, C.E.; Wang, J.; Tewari, A.K.; Eroshkin, A.; Cho, Y.J.; Witt, H.; Korshunov, A.; Read, T.A.; Sun, J.L.; et al. An animal model of MYC-driven medulloblastoma. Cancer Cell 2012, 21, 155–167.
- MacDonald, T.J.; Rood, B.R.; Santi, M.R.; Vezina, G.; Bingaman, K.; Cogen, P.H.; Packer, R.J. Advances in the diagnosis, molecular genetics, and treatment of pediatric embryonal CNS tumors. Oncologist 2003, 8, 174–186.
- Zhou, M.; Jiang, J. Gli Phosphorylation Code in Hedgehog Signal Transduction. Front. Cell Dev. Biol. 2022, 10, 846927.
- Zhang, Q.; Jiang, J. Regulation of Hedgehog Signal Transduction by Ubiquitination and Deubiquitination. Int. J. Mol. Sci. 2021, 22, 13338.
- Canettieri, G.; Di Marcotullio, L.; Greco, A.; Coni, S.; Antonucci, L.; Infante, P.; Pietrosanti, L.; De Smaele, E.; Ferretti, E.; Miele, E.; et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat. Cell Biol. 2010, 12, 132–142.
- Blanc, R.S.; Richard, S. Arginine Methylation: The Coming of Age. Mol. Cell 2017, 65, 8–24.
- Yang, Y.; Bedford, M.T. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 2013, 13, 37–50.
- Smith, E.; Zhou, W.; Shindiapina, P.; Sif, S.; Li, C.; Baiocchi, R.A. Recent advances in targeting protein arginine methyltransferase enzymes in cancer therapy. Expert. Opin. Ther. Targets 2018, 22, 527–545.
- Wolf, S.S. The protein arginine methyltransferase family: An update about function, new perspectives and the physiological role in humans. Cell Mol. Life Sci. 2009, 66, 2109–2121.
- Musiani, D.; Bok, J.; Massignani, E.; Wu, L.; Tabaglio, T.; Ippolito, M.R.; Cuomo, A.; Ozbek, U.; Zorgati, H.; Ghoshdastider, U.; et al. Proteomics profiling of arginine methylation defines PRMT5 substrate specificity. Sci. Signal. 2019, 12, eaat8388.
- Abe, Y.; Suzuki, Y.; Kawamura, K.; Tanaka, N. MEP50/PRMT5-mediated methylation activates GLI1 in Hedgehog signalling through inhibition of ubiquitination by the ITCH/NUMB complex. Commun. Biol. 2019, 2, 23.
- Chaturvedi, N.K.; Mahapatra, S.; Kesherwani, V.; Kling, M.J.; Shukla, M.; Ray, S.; Kanchan, R.; Perumal, N.; McGuire, T.R.; Sharp, J.G.; et al. Role of protein arginine methyltransferase 5 in group 3 (MYC-driven) Medulloblastoma. BMC Cancer 2019, 19, 1056.
- Wynn, D.T.; Rodriguez-Blanco, J.; Long, J.; Yang, F.; Shen, C.; Fei, D.; Tang, H.Y.; Hanson, D.; Robbins, D.J. Protein arginine methyltransferase 5 regulates SHH-subgroup medulloblastoma progression. Neurooncol. Adv. 2022, 4, vdac144.
- Pal, S.; Baiocchi, R.A.; Byrd, J.C.; Grever, M.R.; Jacob, S.T.; Sif, S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007, 26, 3558–3569.
- Wang, L.; Pal, S.; Sif, S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol. Cell Biol. 2008, 28, 6262–6277.
- Koh, C.M.; Bezzi, M.; Low, D.H.; Ang, W.X.; Teo, S.X.; Gay, F.P.; Al-Haddawi, M.; Tan, S.Y.; Osato, M.; Sabo, A.; et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 2015, 523, 96–100.
- Chan-Penebre, E.; Kuplast, K.G.; Majer, C.R.; Boriack-Sjodin, P.A.; Wigle, T.J.; Johnston, L.D.; Rioux, N.; Munchhof, M.J.; Jin, L.; Jacques, S.L.; et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol. 2015, 11, 432–437.
- Brehmer, D.; Beke, L.; Wu, T.; Millar, H.J.; Moy, C.; Sun, W.; Mannens, G.; Pande, V.; Boeckx, A.; van Heerde, E.; et al. Discovery and Pharmacological Characterization of JNJ-64619178, a Novel Small-Molecule Inhibitor of PRMT5 with Potent Antitumor Activity. Mol. Cancer Ther. 2021, 20, 2317–2328.
- Litzler, L.C.; Zahn, A.; Meli, A.P.; Hebert, S.; Patenaude, A.M.; Methot, S.P.; Sprumont, A.; Bois, T.; Kitamura, D.; Costantino, S.; et al. PRMT5 is essential for B cell development and germinal center dynamics. Nat. Commun. 2019, 10, 22.
- Barczak, W.; Jin, L.; Carr, S.M.; Munro, S.; Ward, S.; Kanapin, A.; Samsonova, A.; La Thangue, N.B. PRMT5 promotes cancer cell migration and invasion through the E2F pathway. Cell Death Dis. 2020, 11, 572.
- Guderian, G.; Peter, C.; Wiesner, J.; Sickmann, A.; Schulze-Osthoff, K.; Fischer, U.; Grimmler, M. RioK1, a new interactor of protein arginine methyltransferase 5 (PRMT5), competes with pICln for binding and modulates PRMT5 complex composition and substrate specificity. J. Biol. Chem. 2011, 286, 1976–1986.
- Krzyzanowski, A.; Gasper, R.; Adihou, H.; Hart, P.; Waldmann, H. Biochemical Investigation of the Interaction of pICln, RioK1 and COPR5 with the PRMT5-MEP50 Complex. Chembiochem 2021, 22, 1908–1914.
- Mulvaney, K.M.; Blomquist, C.; Acharya, N.; Li, R.; Ranaghan, M.J.; O’Keefe, M.; Rodriguez, D.J.; Young, M.J.; Kesar, D.; Pal, D.; et al. Molecular basis for substrate recruitment to the PRMT5 methylosome. Mol. Cell 2021, 81, 3481–3495.e7.
- Fu, S.; Zheng, Q.; Zhang, D.; Lin, C.; Ouyang, L.; Zhang, J.; Chen, L. Medicinal chemistry strategies targeting PRMT5 for cancer therapy. Eur. J. Med. Chem. 2022, 244, 114842.
- Burgos, E.S.; Wilczek, C.; Onikubo, T.; Bonanno, J.B.; Jansong, J.; Reimer, U.; Shechter, D. Histone H2A and H4 N-terminal tails are positioned by the MEP50 WD repeat protein for efficient methylation by the PRMT5 arginine methyltransferase. J. Biol. Chem. 2015, 290, 9674–9689.
- Scoumanne, A.; Zhang, J.; Chen, X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res. 2009, 37, 4965–4976.
- Wei, H.; Mundade, R.; Lange, K.C.; Lu, T. Protein arginine methylation of non-histone proteins and its role in diseases. Cell Cycle 2014, 13, 32–41.
- Stopa, N.; Krebs, J.E.; Shechter, D. The PRMT5 arginine methyltransferase: Many roles in development, cancer and beyond. Cell Mol. Life Sci. 2015, 72, 2041–2059.
- Richard, S.; Morel, M.; Cleroux, P. Arginine methylation regulates IL-2 gene expression: A role for protein arginine methyltransferase 5 (PRMT5). Biochem. J. 2005, 388, 379–386.
- Zhou, Z.; Sun, X.; Zou, Z.; Sun, L.; Zhang, T.; Guo, S.; Wen, Y.; Liu, L.; Wang, Y.; Qin, J.; et al. PRMT5 regulates Golgi apparatus structure through methylation of the golgin GM130. Cell Res. 2010, 20, 1023–1033.
- Hsu, J.M.; Chen, C.T.; Chou, C.K.; Kuo, H.P.; Li, L.Y.; Lin, C.Y.; Lee, H.J.; Wang, Y.N.; Liu, M.; Liao, H.W.; et al. Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nat. Cell Biol. 2011, 13, 174–181.
- Karkhanis, V.; Hu, Y.J.; Baiocchi, R.A.; Imbalzano, A.N.; Sif, S. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem. Sci. 2011, 36, 633–641.
- Bandyopadhyay, S.; Harris, D.P.; Adams, G.N.; Lause, G.E.; McHugh, A.; Tillmaand, E.G.; Money, A.; Willard, B.; Fox, P.L.; Dicorleto, P.E. HOXA9 methylation by PRMT5 is essential for endothelial cell expression of leukocyte adhesion molecules. Mol. Cell Biol. 2012, 32, 1202–1213.
- Cho, E.C.; Zheng, S.; Munro, S.; Liu, G.; Carr, S.M.; Moehlenbrink, J.; Lu, Y.C.; Stimson, L.; Khan, O.; Konietzny, R.; et al. Arginine methylation controls growth regulation by E2F-1. EMBO J. 2012, 31, 1785–1797.
- Chen, M.; Yi, B.; Sun, J. Inhibition of cardiomyocyte hypertrophy by protein arginine methyltransferase 5. J. Biol. Chem. 2014, 289, 24325–24335.
- Fay, M.M.; Clegg, J.M.; Uchida, K.A.; Powers, M.A.; Ullman, K.S. Enhanced arginine methylation of programmed cell death 4 protein during nutrient deprivation promotes tumor cell viability. J. Biol. Chem. 2014, 289, 17541–17552.
- Liu, L.; Zhao, X.; Zhao, L.; Li, J.; Yang, H.; Zhu, Z.; Liu, J.; Huang, G. Arginine Methylation of SREBP1a via PRMT5 Promotes De Novo Lipogenesis and Tumor Growth. Cancer Res. 2016, 76, 1260–1272.
- Zhao, D.Y.; Gish, G.; Braunschweig, U.; Li, Y.; Ni, Z.; Schmitges, F.W.; Zhong, G.; Liu, K.; Li, W.; Moffat, J.; et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature 2016, 529, 48–53.
- Fong, J.Y.; Pignata, L.; Goy, P.A.; Kawabata, K.C.; Lee, S.C.; Koh, C.M.; Musiani, D.; Massignani, E.; Kotini, A.G.; Penson, A.; et al. Therapeutic Targeting of RNA Splicing Catalysis through Inhibition of Protein Arginine Methylation. Cancer Cell 2019, 36, 194–209.e199.
- Checa-Rodriguez, C.; Cepeda-Garcia, C.; Ramon, J.; Lopez-Saavedra, A.; Balestra, F.R.; Dominguez-Sanchez, M.S.; Gomez-Cabello, D.; Huertas, P. Methylation of the central transcriptional regulator KLF4 by PRMT5 is required for DNA end resection and recombination. DNA Repair 2020, 94, 102902.
- Park, J.H.; Szemes, M.; Vieira, G.C.; Melegh, Z.; Malik, S.; Heesom, K.J.; Von Wallwitz-Freitas, L.; Greenhough, A.; Brown, K.W.; Zheng, Y.G.; et al. Protein arginine methyltransferase 5 is a key regulator of the MYCN oncoprotein in neuroblastoma cells. Mol. Oncol. 2015, 9, 617–627.
- Wei, H.; Wang, B.; Miyagi, M.; She, Y.; Gopalan, B.; Huang, D.B.; Ghosh, G.; Stark, G.R.; Lu, T. PRMT5 dimethylates R30 of the p65 subunit to activate NF-kappaB. Proc. Natl. Acad. Sci. USA 2013, 110, 13516–13521.
- Tee, W.W.; Pardo, M.; Theunissen, T.W.; Yu, L.; Choudhary, J.S.; Hajkova, P.; Surani, M.A. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010, 24, 2772–2777.
- Gkountela, S.; Li, Z.; Chin, C.J.; Lee, S.A.; Clark, A.T. PRMT5 is required for human embryonic stem cell proliferation but not pluripotency. Stem Cell Rev. Rep. 2014, 10, 230–239.
- Bezzi, M.; Teo, S.X.; Muller, J.; Mok, W.C.; Sahu, S.K.; Vardy, L.A.; Bonday, Z.Q.; Guccione, E. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 2013, 27, 1903–1916.
- Dacwag, C.S.; Bedford, M.T.; Sif, S.; Imbalzano, A.N. Distinct protein arginine methyltransferases promote ATP-dependent chromatin remodeling function at different stages of skeletal muscle differentiation. Mol. Cell Biol. 2009, 29, 1909–1921.
- Li, Z.; Yu, J.; Hosohama, L.; Nee, K.; Gkountela, S.; Chaudhari, S.; Cass, A.A.; Xiao, X.; Clark, A.T. The Sm protein methyltransferase PRMT5 is not required for primordial germ cell specification in mice. EMBO J. 2015, 34, 748–758.
- Zhang, T.; Gunther, S.; Looso, M.; Kunne, C.; Kruger, M.; Kim, J.; Zhou, Y.; Braun, T. Prmt5 is a regulator of muscle stem cell expansion in adult mice. Nat. Commun. 2015, 6, 7140.
- Shailesh, H.; Zakaria, Z.Z.; Baiocchi, R.; Sif, S. Protein arginine methyltransferase 5 (PRMT5) dysregulation in cancer. Oncotarget 2018, 9, 36705–36718.
- Pal, S.; Vishwanath, S.N.; Erdjument-Bromage, H.; Tempst, P.; Sif, S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell Biol. 2004, 24, 9630–9645.
- Mongiardi, M.P.; Savino, M.; Bartoli, L.; Beji, S.; Nanni, S.; Scagnoli, F.; Falchetti, M.L.; Favia, A.; Farsetti, A.; Levi, A.; et al. Myc and Omomyc functionally associate with the Protein Arginine Methyltransferase 5 (PRMT5) in glioblastoma cells. Sci. Rep. 2015, 5, 15494.
- Yan, F.; Alinari, L.; Lustberg, M.E.; Martin, L.K.; Cordero-Nieves, H.M.; Banasavadi-Siddegowda, Y.; Virk, S.; Barnholtz-Sloan, J.; Bell, E.H.; Wojton, J.; et al. Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Res. 2014, 74, 1752–1765.
- Han, X.; Li, R.; Zhang, W.; Yang, X.; Wheeler, C.G.; Friedman, G.K.; Province, P.; Ding, Q.; You, Z.; Fathallah-Shaykh, H.M.; et al. Expression of PRMT5 correlates with malignant grade in gliomas and plays a pivotal role in tumor growth in vitro. J. Neurooncol. 2014, 118, 61–72.
- Wang, Y.; Hu, W.; Yuan, Y. Protein Arginine Methyltransferase 5 (PRMT5) as an Anticancer Target and Its Inhibitor Discovery. J. Med. Chem. 2018, 61, 9429–9441.
- Ibrahim, R.; Matsubara, D.; Osman, W.; Morikawa, T.; Goto, A.; Morita, S.; Ishikawa, S.; Aburatani, H.; Takai, D.; Nakajima, J.; et al. Expression of PRMT5 in lung adenocarcinoma and its significance in epithelial-mesenchymal transition. Hum. Pathol. 2014, 45, 1397–1405.
- Gu, Z.; Li, Y.; Lee, P.; Liu, T.; Wan, C.; Wang, Z. Protein arginine methyltransferase 5 functions in opposite ways in the cytoplasm and nucleus of prostate cancer cells. PLoS ONE 2012, 7, e44033.
- Nicholas, C.; Yang, J.; Peters, S.B.; Bill, M.A.; Baiocchi, R.A.; Yan, F.; Sif, S.; Tae, S.; Gaudio, E.; Wu, X.; et al. PRMT5 is upregulated in malignant and metastatic melanoma and regulates expression of MITF and p27(Kip1). PLoS ONE 2013, 8, e74710.
- Koh, C.M.; Bezzi, M.; Guccion, E. The Where and the How of PRMT5. Curr. Mol. Bio Rep. 2015, 1, 19–28.
- Chitiprolu, M.; Jagow, C.; Tremblay, V.; Bondy-Chorney, E.; Paris, G.; Savard, A.; Palidwor, G.; Barry, F.A.; Zinman, L.; Keith, J.; et al. A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy. Nat. Commun. 2018, 9, 2794.
- Quan, X.; Yue, W.; Luo, Y.; Cao, J.; Wang, H.; Wang, Y.; Lu, Z. The protein arginine methyltransferase PRMT5 regulates Abeta-induced toxicity in human cells and Caenorhabditis elegans models of Alzheimer’s disease. J. Neurochem. 2015, 134, 969–977.
- Ratovitski, T.; Arbez, N.; Stewart, J.C.; Chighladze, E.; Ross, C.A. PRMT5- mediated symmetric arginine dimethylation is attenuated by mutant huntingtin and is impaired in Huntington’s disease (HD). Cell Cycle 2015, 14, 1716–1729.
- Banasavadi-Siddegowda, Y.K.; Russell, L.; Frair, E.; Karkhanis, V.A.; Relation, T.; Yoo, J.Y.; Zhang, J.; Sif, S.; Imitola, J.; Baiocchi, R.; et al. PRMT5-PTEN molecular pathway regulates senescence and self-renewal of primary glioblastoma neurosphere cells. Oncogene 2017, 36, 263–274.
- Braun, C.J.; Stanciu, M.; Boutz, P.L.; Patterson, J.C.; Calligaris, D.; Higuchi, F.; Neupane, R.; Fenoglio, S.; Cahill, D.P.; Wakimoto, H.; et al. Coordinated Splicing of Regulatory Detained Introns within Oncogenic Transcripts Creates an Exploitable Vulnerability in Malignant Glioma. Cancer Cell 2017, 32, 411–426.e411.
- Sachamitr, P.; Ho, J.C.; Ciamponi, F.E.; Ba-Alawi, W.; Coutinho, F.J.; Guilhamon, P.; Kushida, M.M.; Cavalli, F.M.G.; Lee, L.; Rastegar, N.; et al. PRMT5 inhibition disrupts splicing and stemness in glioblastoma. Nat. Commun. 2021, 12, 979.
- Holmes, B.; Benavides-Serrato, A.; Saunders, J.T.; Landon, K.A.; Schreck, A.J.; Nishimura, R.N.; Gera, J. The protein arginine methyltransferase PRMT5 confers therapeutic resistance to mTOR inhibition in glioblastoma. J. Neurooncol. 2019, 145, 11–22.
- Du, C.; Hansen, L.J.; Singh, S.X.; Wang, F.; Sun, R.; Moure, C.J.; Roso, K.; Greer, P.K.; Yan, H.; He, Y. A PRMT5-RNF168-SMURF2 Axis Controls H2AX Proteostasis. Cell Rep. 2019, 28, 3199–3211.e3195.
- Huang, L.; Zhang, X.O.; Rozen, E.J.; Sun, X.; Sallis, B.; Verdejo-Torres, O.; Wigglesworth, K.; Moon, D.; Huang, T.; Cavaretta, J.P.; et al. PRMT5 activates AKT via methylation to promote tumor metastasis. Nat. Commun. 2022, 13, 3955.
- Chen, H.; Lorton, B.; Gupta, V.; Shechter, D. A TGFbeta-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene 2017, 36, 373–386.
- Fu, T.; Lv, X.; Kong, Q.; Yuan, C. A novel SHARPIN-PRMT5-H3R2me1 axis is essential for lung cancer cell invasion. Oncotarget 2017, 8, 54809–54820.
- Jing, P.; Zhao, N.; Ye, M.; Zhang, Y.; Zhang, Z.; Sun, J.; Wang, Z.; Zhang, J.; Gu, Z. Protein arginine methyltransferase 5 promotes lung cancer metastasis via the epigenetic regulation of miR-99 family/FGFR3 signaling. Cancer Lett. 2018, 427, 38–48.
- Zhou, H.; Chang, J.; Zhang, J.; Zheng, H.; Miao, X.; Mo, H.; Sun, J.; Jia, Q.; Qi, G. PRMT5 activates KLF5 by methylation to facilitate lung cancer. J. Cell Mol. Med. 2023, 1–15.
- Jiang, H.; Zhu, Y.; Zhou, Z.; Xu, J.; Jin, S.; Xu, K.; Zhang, H.; Sun, Q.; Wang, J.; Xu, J. PRMT5 promotes cell proliferation by inhibiting BTG2 expression via the ERK signaling pathway in hepatocellular carcinoma. Cancer Med. 2018, 7, 869–882.
- Li, Z.; Zhang, J.; Liu, X.; Li, S.; Wang, Q.; Di, C.; Hu, Z.; Yu, T.; Ding, J.; Li, J.; et al. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat. Commun. 2018, 9, 1572.
- Zhu, K.; Peng, Y.; Hu, J.; Zhan, H.; Yang, L.; Gao, Q.; Jia, H.; Luo, R.; Dai, Z.; Tang, Z.; et al. Metadherin-PRMT5 complex enhances the metastasis of hepatocellular carcinoma through the WNT-beta-catenin signaling pathway. Carcinogenesis 2020, 41, 130–138.
- Hing, Z.A.; Walker, J.S.; Whipp, E.C.; Brinton, L.; Cannon, M.; Zhang, P.; Sher, S.; Cempre, C.B.; Brown, F.; Smith, P.L.; et al. Dysregulation of PRMT5 in chronic lymphocytic leukemia promotes progression with high risk of Richter’s transformation. Nat. Commun. 2023, 14, 97.
- Qin, Y.; Hu, Q.; Xu, J.; Ji, S.; Dai, W.; Liu, W.; Xu, W.; Sun, Q.; Zhang, Z.; Ni, Q.; et al. PRMT5 enhances tumorigenicity and glycolysis in pancreatic cancer via the FBW7/cMyc axis. Cell Commun. Signal. 2019, 17, 30.
- Orben, F.; Lankes, K.; Schneeweis, C.; Hassan, Z.; Jakubowsky, H.; Krauss, L.; Boniolo, F.; Schneider, C.; Schafer, A.; Murr, J.; et al. Epigenetic drug screening defines a PRMT5 inhibitor-sensitive pancreatic cancer subtype. JCI Insight 2022, 7, e151353.
- Dong, Y.; Song, C.; Wang, Y.; Lei, Z.; Xu, F.; Guan, H.; Chen, A.; Li, F. Inhibition of PRMT5 suppresses osteoclast differentiation and partially protects against ovariectomy-induced bone loss through downregulation of CXCL10 and RSAD2. Cell Signal. 2017, 34, 55–65.
- Kota, S.K.; Roening, C.; Patel, N.; Kota, S.B.; Baron, R. PRMT5 inhibition promotes osteogenic differentiation of mesenchymal stromal cells and represses basal interferon stimulated gene expression. Bone 2018, 117, 37–46.
- Mounir, Z.; Korn, J.M.; Westerling, T.; Lin, F.; Kirby, C.A.; Schirle, M.; McAllister, G.; Hoffman, G.; Ramadan, N.; Hartung, A.; et al. ERG signaling in prostate cancer is driven through PRMT5-dependent methylation of the Androgen Receptor. eLife 2016, 5, e13964.
- Beketova, E.; Fang, S.; Owens, J.L.; Liu, S.; Chen, X.; Zhang, Q.; Asberry, A.M.; Deng, X.; Malola, J.; Huang, J.; et al. Protein Arginine Methyltransferase 5 Promotes pICln-Dependent Androgen Receptor Transcription in Castration-Resistant Prostate Cancer. Cancer Res. 2020, 80, 4904–4917.
- Bao, X.; Zhao, S.; Liu, T.; Liu, Y.; Liu, Y.; Yang, X. Overexpression of PRMT5 promotes tumor cell growth and is associated with poor disease prognosis in epithelial ovarian cancer. J. Histochem. Cytochem. 2013, 61, 206–217.
- Wang, Z.; Kong, J.; Wu, Y.; Zhang, J.; Wang, T.; Li, N.; Fan, J.; Wang, H.; Zhang, J.; Ling, R. PRMT5 determines the sensitivity to chemotherapeutics by governing stemness in breast cancer. Breast Cancer Res. Treat. 2018, 168, 531–542.
- Chiang, K.; Zielinska, A.E.; Shaaban, A.M.; Sanchez-Bailon, M.P.; Jarrold, J.; Clarke, T.L.; Zhang, J.; Francis, A.; Jones, L.J.; Smith, S.; et al. PRMT5 Is a Critical Regulator of Breast Cancer Stem Cell Function via Histone Methylation and FOXP1 Expression. Cell Rep. 2017, 21, 3498–3513.
- Yin, S.; Liu, L.; Brobbey, C.; Palanisamy, V.; Ball, L.E.; Olsen, S.K.; Ostrowski, M.C.; Gan, W. PRMT5-mediated arginine methylation activates AKT kinase to govern tumorigenesis. Nat. Commun. 2021, 12, 3444.
- Hu, D.; Gur, M.; Zhou, Z.; Gamper, A.; Hung, M.C.; Fujita, N.; Lan, L.; Bahar, I.; Wan, Y. Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat. Commun. 2015, 6, 8419.
- Rengasamy, M.; Zhang, F.; Vashisht, A.; Song, W.M.; Aguilo, F.; Sun, Y.; Li, S.; Zhang, W.; Zhang, B.; Wohlschlegel, J.A.; et al. The PRMT5/WDR77 complex regulates alternative splicing through ZNF326 in breast cancer. Nucleic Acids Res. 2017, 45, 11106–11120.
- Powers, M.A.; Fay, M.M.; Factor, R.E.; Welm, A.L.; Ullman, K.S. Protein arginine methyltransferase 5 accelerates tumor growth by arginine methylation of the tumor suppressor programmed cell death 4. Cancer Res. 2011, 71, 5579–5587.
- Yi, J.; Wu, J. Epigenetic regulation in medulloblastoma. Mol. Cell Neurosci. 2018, 87, 65–76.
- Roussel, M.F.; Stripay, J.L. Epigenetic Drivers in Pediatric Medulloblastoma. Cerebellum 2018, 17, 28–36.
- Dubuc, A.M.; Remke, M.; Korshunov, A.; Northcott, P.A.; Zhan, S.H.; Mendez-Lago, M.; Kool, M.; Jones, D.T.; Unterberger, A.; Morrissy, A.S.; et al. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol. 2013, 125, 373–384.
- Alimova, I.; Venkataraman, S.; Harris, P.; Marquez, V.E.; Northcott, P.A.; Dubuc, A.; Taylor, M.D.; Foreman, N.K.; Vibhakar, R. Targeting the enhancer of zeste homologue 2 in medulloblastoma. Int. J. Cancer 2012, 131, 1800–1809.
- Favia, A.; Salvatori, L.; Nanni, S.; Iwamoto-Stohl, L.K.; Valente, S.; Mai, A.; Scagnoli, F.; Fontanella, R.A.; Totta, P.; Nasi, S.; et al. The Protein Arginine Methyltransferases 1 and 5 affect Myc properties in glioblastoma stem cells. Sci. Rep. 2019, 9, 15925.
- Chittka, A.; Nitarska, J.; Grazini, U.; Richardson, W.D. Transcription factor positive regulatory domain 4 (PRDM4) recruits protein arginine methyltransferase 5 (PRMT5) to mediate histone arginine methylation and control neural stem cell proliferation and differentiation. J. Biol. Chem. 2012, 287, 42995–43006.
- Zhou, Z.; Feng, Z.; Hu, D.; Yang, P.; Gur, M.; Bahar, I.; Cristofanilli, M.; Gradishar, W.J.; Xie, X.Q.; Wan, Y. A novel small-molecule antagonizes PRMT5-mediated KLF4 methylation for targeted therapy. EBioMedicine 2019, 44, 98–111.
- Wu, Q.; Schapira, M.; Arrowsmith, C.H.; Barsyte-Lovejoy, D. Protein arginine methylation: From enigmatic functions to therapeutic targeting. Nat. Rev. Drug Discov. 2021, 20, 509–530.
- Feustel, K.; Falchook, G.S. Protein Arginine Methyltransferase 5 (PRMT5) Inhibitors in Oncology Clinical Trials: A review. J. Immunother. Precis. Oncol. 2022, 5, 58–67.
- Millar, H.J.; Brehmer, D.; Verhulst, T.; Haddish-Berhane, N.; Greway, T.; Gaffney, D.; Boeckx, A.; Heerde, E.V.; Nys, T.; Portale, J.; et al. In vivo efficacy and pharmacodynamic modulation of JNJ-64619178, a selective PRMT5 inhibitor, in human lung and hematologic preclinical models . In Proceedings of the American Association for Cancer Research Annual Meeting 2019, Atlanta, GA, USA, 29 March–3 April 2019; AACR: Philadelphia, PA, USA, 2019. Volume 79 (Suppl 13), Abstract nr 950.
- Jensen-Pergakes, K.; Tatlock, J.; Maegley, K.A.; McAlpine, I.J.; McTigue, M.; Xie, T.; Dillon, C.P.; Wang, Y.; Yamazaki, S.; Spiegel, N.; et al. SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance. Mol. Cancer Ther. 2022, 21, 3–15.
- Duncan, K.W.; Rioux, N.; Boriack-Sjodin, P.A.; Munchhof, M.J.; Reiter, L.A.; Majer, C.R.; Jin, L.; Johnston, L.D.; Chan-Penebre, E.; Kuplast, K.G.; et al. Structure and Property Guided Design in the Identification of PRMT5 Tool Compound EPZ015666. ACS Med. Chem. Lett. 2016, 7, 162–166.
- Gulla, A.; Hideshima, T.; Bianchi, G.; Fulciniti, M.; Kemal Samur, M.; Qi, J.; Tai, Y.T.; Harada, T.; Morelli, E.; Amodio, N.; et al. Protein arginine methyltransferase 5 has prognostic relevance and is a druggable target in multiple myeloma. Leukemia 2018, 32, 996–1002.
- Hu, G.; Wang, X.; Han, Y.; Wang, P. Protein arginine methyltransferase 5 promotes bladder cancer growth through inhibiting NF-kB dependent apoptosis. EXCLI J. 2018, 17, 1157–1166.
- Kaushik, S.; Liu, F.; Veazey, K.J.; Gao, G.; Das, P.; Neves, L.F.; Lin, K.; Zhong, Y.; Lu, Y.; Giuliani, V.; et al. Genetic deletion or small-molecule inhibition of the arginine methyltransferase PRMT5 exhibit anti-tumoral activity in mouse models of MLL-rearranged AML. Leukemia 2018, 32, 499–509.
- Serio, J.; Ropa, J.; Chen, W.; Mysliwski, M.; Saha, N.; Chen, L.; Wang, J.; Miao, H.; Cierpicki, T.; Grembecka, J.; et al. The PAF complex regulation of Prmt5 facilitates the progression and maintenance of MLL fusion leukemia. Oncogene 2018, 37, 450–460.
- AbuHammad, S.; Cullinane, C.; Martin, C.; Bacolas, Z.; Ward, T.; Chen, H.; Slater, A.; Ardley, K.; Kirby, L.; Chan, K.T.; et al. Regulation of PRMT5-MDM4 axis is critical in the response to CDK4/6 inhibitors in melanoma. Proc. Natl. Acad. Sci. USA 2019, 116, 17990–18000.
- Gerhart, S.V.; Kellner, W.A.; Thompson, C.; Pappalardi, M.B.; Zhang, X.P.; Montes de Oca, R.; Penebre, E.; Duncan, K.; Boriack-Sjodin, A.; Le, B.; et al. Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing. Sci. Rep. 2018, 8, 9711.
- Vinet, M.; Suresh, S.; Maire, V.; Monchecourt, C.; Nemati, F.; Lesage, L.; Pierre, F.; Ye, M.; Lescure, A.; Brisson, A.; et al. Protein arginine methyltransferase 5: A novel therapeutic target for triple-negative breast cancers. Cancer Med. 2019, 8, 2414–2428.
- Zhu, F.; Guo, H.; Bates, P.D.; Zhang, S.; Zhang, H.; Nomie, K.J.; Li, Y.; Lu, L.; Seibold, K.R.; Wang, F.; et al. PRMT5 is upregulated by B-cell receptor signaling and forms a positive-feedback loop with PI3K/AKT in lymphoma cells. Leukemia 2019, 33, 2898–2911.
- Villalona-Calero, M.A.; Patnaik, A.; Maki, R.G.; O’Neil, B.; Abbruzzese, J.L.; Dagogo-Jack, I.; Devarakonda, S.; Wahlroos, S.; Lin, C.-C.; Fujiwara, Y.; et al. Design and rationale of a phase 1 dose-escalation study of AMG 193, a methylthioadenosine (MTA)-cooperative PRMT5 inhibitor, in patients with advanced methylthioadenosine phosphorylase (MTAP)-null solid tumors. J. Clin. Oncol. 2022, 40, TPS3167.
- Bhagwat, N.; Zhang, Y.; Lin, H.; Wang, M.; Rominger, D.; Emm, T.; Chugani-Mahtani, D.; Angelis, D.; Shetty, R.; Leal, R.; et al. Preclinical Characterization of PRT543, a Potent and Selective Inhibitor of Protein Arginine Methyltransferase 5 (PRMT5), with Broad Antitumor Activity in In vitro and In vivo Models. In Proceedings of the Annual Meeting of the American Association for Cancer Research, Philadelphia, PA, USA, 27–28 April 2020.
- Falchook, G. A phase 1 dose-escalation study of protein arginine methyltransferase 5 (PRMT5) brain-penetrant inhibitor PRT811 in patients with advanced solid tumors, including recurrent high-grade gliomas. In Proceedings of the AACR-NCI-EORTC Virtual Conference, Virtual, 7–10 October 2021; Available online: www.aacr.org/meeting/aacr-nci-eortc-international (accessed on 17 November 2023).
- Zhang, M.; Tsai, A.; Cottrell, K.M.; Haines, B.B.; Wilker, E.; DiBenedetto, H.; Weitzman, R.; Huang, A.; Davis, C.B.; Maxwell, J.P.; et al. TNG908, a brain-penetrant MTA-cooperative PRMT5 inhibitor, is efficacious in preclinical glioblastoma models. In Proceedings of the American Association for Cancer Research Annual Meeting 2023, Orlando, FL, USA, 14–19 April 2023; Part 1 (Regular and Invited Abstracts). AACR: Philadelphia, PA, USA, 2023; Volume 83.
- Vegar, L.; Aranda, R.; Waters, L.; Moya, K.; Hebbert, A.; Smith, C.S.; Hallin, J.; David, B.M.; Engstrom, L.D.; Vanderpool, D.; et al. A novel MTA-cooperative PRMT5 inhibitor, MRTX1719, stabilizes the ternary MTA-PRMT5 complex and leads to synthetic lethality in MTAP deleted cancers . In Proceedings of the American Association for Cancer Research Annual Meeting 2023, Orlando, FL, USA, 14–19 April 2023; Part 1 (Regular and Invited Abstracts). AACR: Philadelphia, PA, USA, 2023; Volume 83.
- Engstrom, L.D.; Aranda, R.; Waters, L.; Moya, K.; Bowcut, V.; Vegar, L.; Trinh, D.; Hebbert, A.; Smith, C.R.; Kulyk, S.; et al. MRTX1719 is an MTA-cooperative PRMT5 inhibitor that exhibits synthetic lethality in preclinical models and patients with MTAP deleted cancer. Cancer Discov. 2023, 13, 2412–2431.
- Bonday, Z.Q.; Cortez, G.S.; Grogan, M.J.; Antonysamy, S.; Weichert, K.; Bocchinfuso, W.P.; Li, F.; Kennedy, S.; Li, B.; Mader, M.M.; et al. LLY-283, a Potent and Selective Inhibitor of Arginine Methyltransferase 5, PRMT5, with Antitumor Activity. ACS Med. Chem. Lett. 2018, 9, 612–617.
- Snyder, K.J.; Zitzer, N.C.; Gao, Y.; Choe, H.K.; Sell, N.E.; Neidemire-Colley, L.; Ignaci, A.; Kale, C.; Devine, R.D.; Abad, M.G.; et al. PRMT5 regulates T cell interferon response and is a target for acute graft-versus-host disease. JCI Insight 2020, 5, e131099.
- Palte, R.L.; Schneider, S.E.; Altman, M.D.; Hayes, R.P.; Kawamura, S.; Lacey, B.M.; Mansueto, M.S.; Reutershan, M.; Siliphaivanh, P.; Sondey, C.; et al. Allosteric Modulation of Protein Arginine Methyltransferase 5 (PRMT5). ACS Med. Chem. Lett. 2020, 11, 1688–1693.
- Sivanandhan, D.; Gajendran, C.; Mohammed, Z.; Gosu, R.; Sher, D.; Mansur, S.; Friedmann-Morvinski, D.; Rajagopal, S.; Rastelli, L. JBI-778, a novel brain-penetrant small molecule PRMT5 inhibitor for treatment of cancer . In Proceedings of the American Association for Cancer Research Annual Meeting 2023, Orlando, FL, USA, 14–19 April 2023; Part 1 (Regular and Invited Abstracts). AACR: Philadelphia, PA, USA, 2023; Volume 83.
- Zheng, J.; Li, B.; Wu, Y.; Wu, X.; Wang, Y. Targeting Arginine Methyltransferase PRMT5 for Cancer Therapy: Updated Progress and Novel Strategies. J. Med. Chem. 2023, 66, 8407–8427.