Plant toxins that target eukaryotic 28S ribosomal RNA (Ribosome Inactivating Proteins, RIPs), such as the catalytic (A) chain of ricin (a dimeric A-B toxin from Ricinus communis) and saporin (a single chain A toxin from Saponaria officinalis), have found widespread use in the construction of immunotoxins. Both these toxins depurinate ribosomal 28S RNA at a specific site (GAGA loop) inhibiting the binding of elongation factor 2 and blocking protein synthesis. Their high toxicity towards eukaryotic cells has thus justified their use as components of chimeric toxins in cancer treatment. A brief description of recombinant RIPs production is given.
- toxin
- immunotoxins
- plant toxin
- recombinant production
1. Saporin SO6
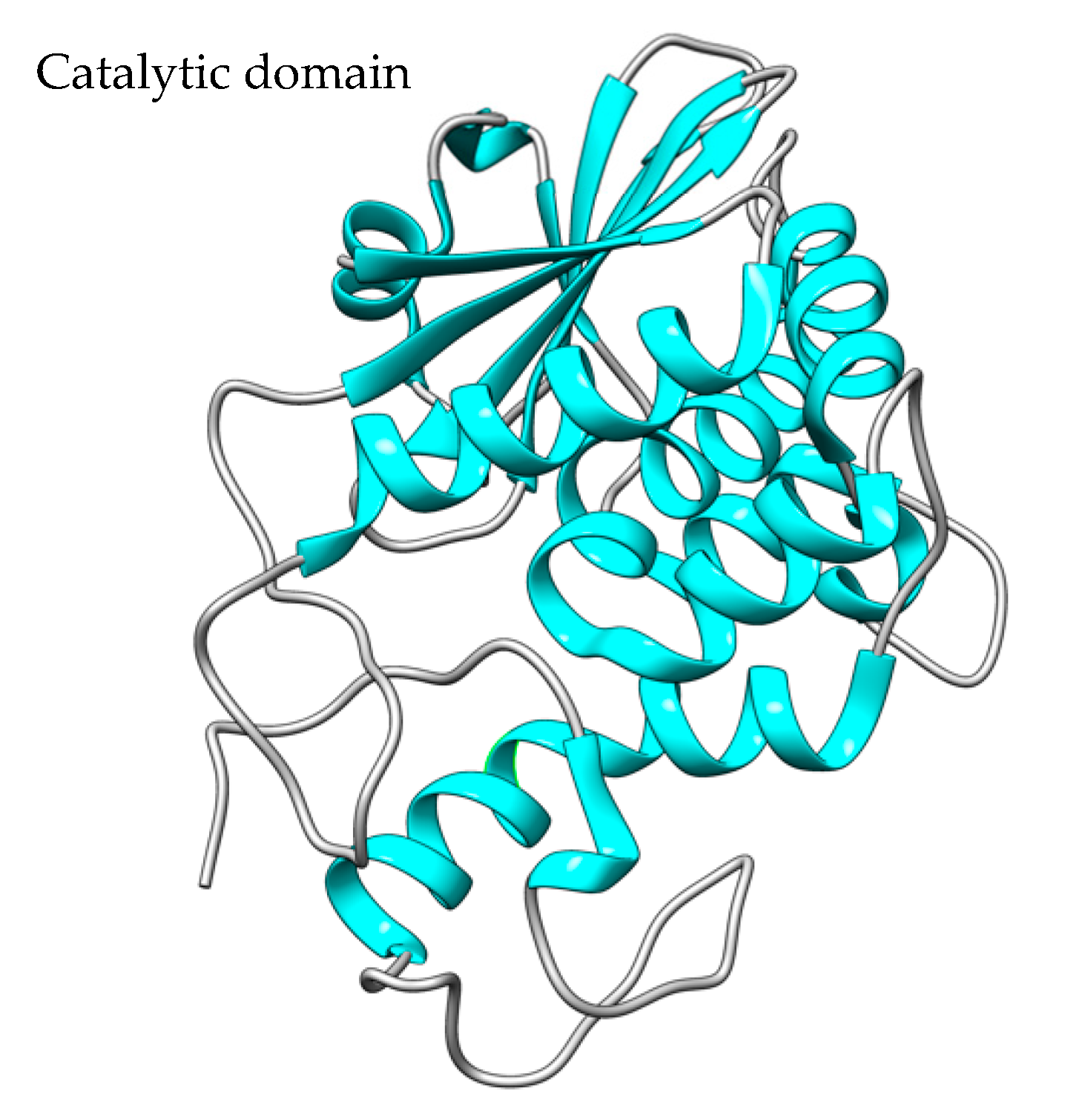
Saporin Expression Hosts
-
Saporin Bacterial expression systems
-
Saporin Yeasts expression systems
-
Saporin expression in Tobacco protoplasts
-
Gene therapy with saporin gene
2. Ricin A Chain (RTA)
RTA Expression Hosts
-
RTA Bacterial expression systems
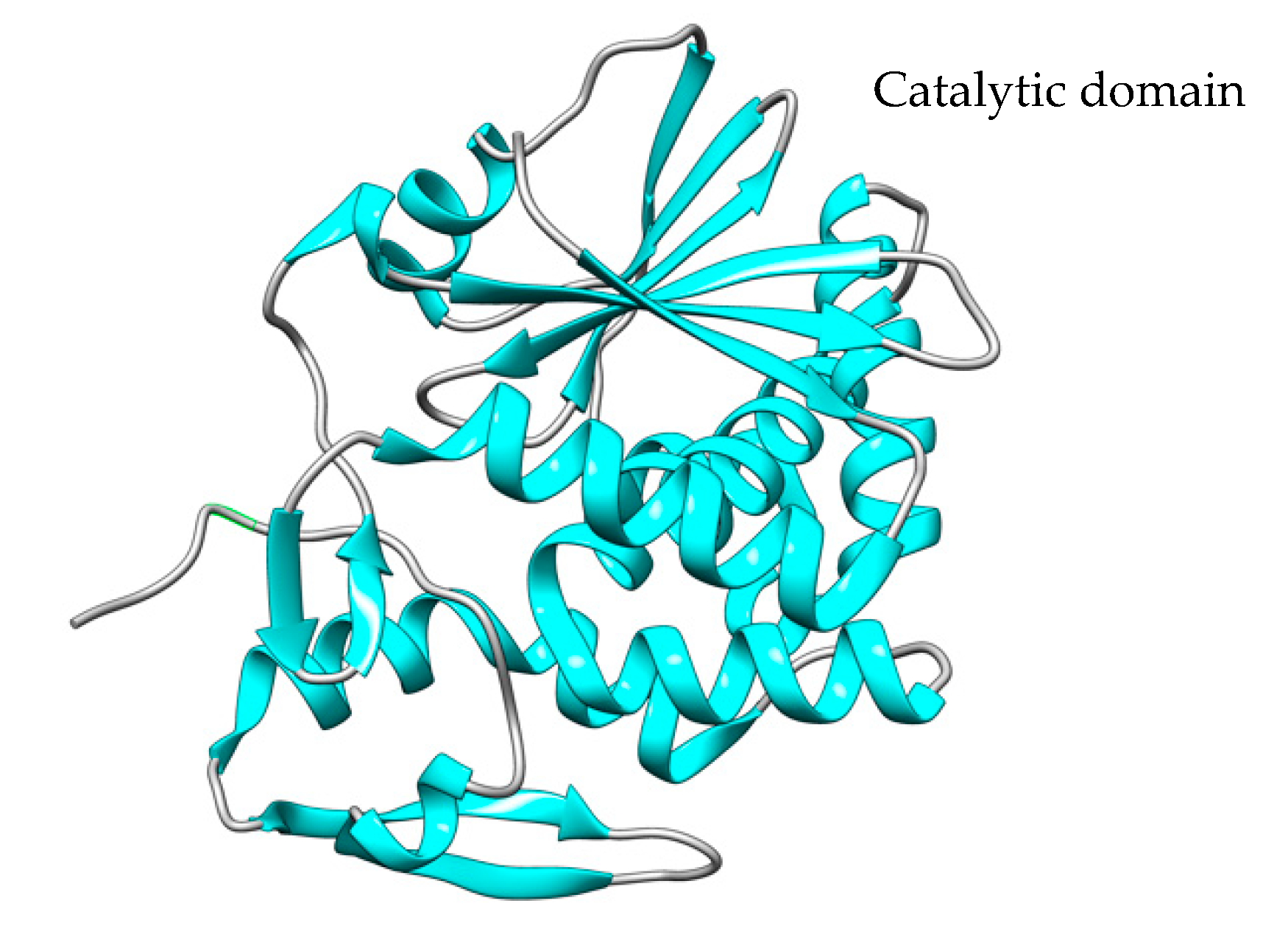
3. Other Plant Toxins
3.1. Bouganin
-
Bacterial expression systems
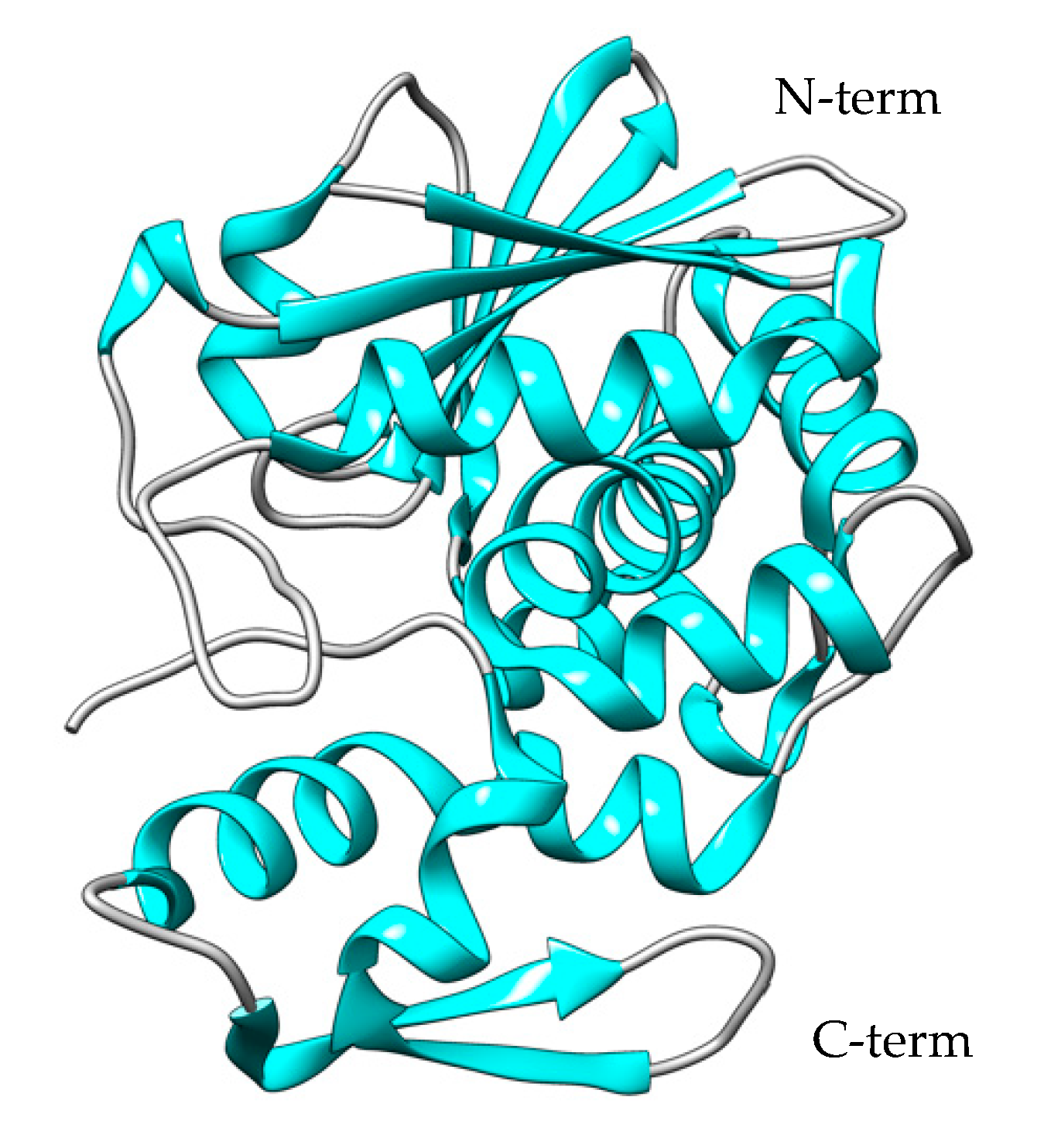
3.2. Pulchellin
This entry is adapted from the peer-reviewed paper 10.3390/toxins15120699
References
- Lappi, D.A.; Esch, F.S.; Barbieri, L.; Stirpe, F.; Soria, M. Characterization of a Saponaria officinalis seed ribosome-inactivating protein: Immunoreactivity and sequence homologies. Biochem. Biophys. Res. Commun. 1985, 129, 934–942.
- Pizzo, E.; Di Maro, A. A new age for biomedical applications of Ribosome Inactivating Proteins (RIPs): From bioconjugate to nanoconstructs. J. Biomed. Sci. 2016, 23, 54.
- Santanché, S.; Bellelli, A.; Brunori, M. The Unusual Stability of Saporin, a Candidate for the Synthesis of Immunotoxins. Biochem. Biophys. Res. Commun. 1997, 234, 129–132.
- Bolognesi, A.; Tazzari, P.L.; Tassi, C.; Gromo, G.; Gobbi, M.; Stirpe, F. A comparison of anti-lymphocyte immunotoxins containing different ribosoma-inactivating proteins and antibodies. Clin. Exp. Immunol. 1992, 89, 341–346.
- Maras, B.; Ippoliti, R.; De Luca, E.; Lendaro, E.; Bellelli, A.; Barra, D.; Bossa, F.; Brunori, M. The amino acid sequence of a ribosome-inactivating protein from Saponaria officinalis seeds. Biochem. Int. 1990, 21, 831–838.
- Endo, Y.; Tsurugi, K. The RNA N-glycosidase activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA. J. Biol. Chem. 1988, 263, 8735–8739.
- Barbieri, L.; Battelli, M.G.; Stirpe, F. Ribosome-inactivating proteins from plants. Biochim. Biophys. Acta 1993, 1154, 237–282.
- Bagga, S.; Seth, D.; Batra, J.K. The cytotoxic activity of ribosome-inactivating protein saporin-6 is attributed to its rRNA N-glycosidase and internucleosomal DNA fragmentation activities. J. Biol. Chem. 2003, 278, 4813–4820.
- Stirpe, F. Ribosome-inactivating proteins: From toxins to useful proteins. Toxicon 2013, 67, 12–16.
- Polito, L.; Bortolotti, M.; Farini, V.; Battelli, M.G.; Barbieri, L.; Bolognesi, A. Saporin induces multiple death pathways in lymphoma cells with different intensity and timing as compared to ricin. Int. J. Biochem. Cell Biol. 2009, 41, 1055–1061.
- Thorpe, P.E.; Brown, A.N.; Bremner, J.A.G., Jr.; Foxwell, B.M.; Stirpe, F. An immunotoxin composed of monoclonal anti-Thy 1.1 antibody and a ribosome-inactivating protein from Saponaria officinalis: Potent antitumor effects in vitro and in vivo. J. Natl. Cancer Inst. 1985, 75, 151–159.
- Glennie, M.J.; McBride, H.M.; Stirpe, F.; Thorpe, P.E.; Worth, A.T.; Stevenson, G.T. Emergence of immunoglobulin variants following treatment of a B cell leukemia with an immunotoxin composed of antiidiotypic antibody and saporin. J. Exp. Med. 1987, 166, 43–62.
- Siena, S.; Lappi, D.A.; Bregni, M.; Formosa, A.; Villa, S.; Soria, M.; Bonadonna, G.; Gianni, A.M. Synthesis and characterization of an antihuman T-lymphocyte saporin immunotoxin (OKT1-SAP) with in vivo stability into nonhuman primates. Blood 1988, 72, 756–765.
- Tazzari, P.L.; Bolognesi, A.; De Totero, D.; Lemoli, R.M.; Fortuna, A.; Conte, R.; Crumpton, M.J.; Stirpe, F. Immunotoxins containing saporin linked to different CD2 monoclonal antibodies: In vitro evaluation. Br. J. Haematol. 1994, 86, 97–105.
- Morland, B.J.; Barley, J.; Boehm, D.; Flavell, S.U.; Ghaleb, N.; Kohler, J.A.; Okayama, K.; Wilkins, B.; Flavell, D.J. Effectiveness of HB2 (anti-CD7)-Saporin immunotoxin in an in vivo model of human T-cell leukaemia developed in severe combined immunodeficient mice. Br. J. Cancer 1994, 69, 279–285.
- Flavell, D.J.; Boehm, D.A.; Okayama, K.; Kohler, J.A.; Flavell, S.U. Therapy of human T-cell acute lymphoblastic leukaemia in severe combined immunodeficient mice with two different anti-CD7-saporin immunotoxins containing hindered or non-hindered disulphide cross-linkers. Int. J. Cancer 1994, 58, 407–414.
- Flavell, D.J.; Flavell, S.U.; Boehm, D.; Emery, L.; Noss, A.; Ling, N.R.; Richardson, P.R.; Hardie, D.; Wright, D.H. Preclinical studies with the anti-CD19-saporin immunotoxin BU12-SAPORIN for the treatment of human-B-cell tumours. Br. J. Cancer 1995, 72, 1373–1379.
- Flavell, D.J.; Boehm, D.A.; Noss, A.M.; Flavell, S.U. Comparison of the potency and therapeutic efficacy of the anti-CD7 immunotoxin HB2-saporin constructed with one or two saporin moieties per immunotoxin molecule. Br. J. Cancer 1997, 75, 1035–1043.
- Flavell, D.J.; Warnes, S.; Noss, A.; Flavell, S.U. Host-mediated antibody-dependent cellular cytotoxicity contributes to the in vivo therapeutic efficacy of an anti-CD7-saporin immunotoxin in a severe combined immunodeficient mouse model of human T-cell acute lymphoblastic leukemia. Cancer Res. 1998, 58, 5787–5794.
- Flavell, D.J.; Warnes, S.L.; Noss, A.L.; Flavell, S.U. Anti-CD7 antibody and immunotoxin treatment of human CD7(+)T-cell leukaemia is significantly less effective in NOD/LtSz-scid mice than in CB.17 scid mice. Br. J. Cancer 2000, 83, 1755–1761.
- Polito, L.; Bortolotti, M.; Mercatelli, D.; Battelli, M.G.; Bolognesi, A. Saporin-S6: A useful tool in cancer therapy. Toxins 2013, 5, 1698–1722.
- Puri, M.; Kaur, I.; Perugini, M.A.; Gupta, R.C. Ribosome-inactivating proteins: Current status and biomedical applications. Drug Discov. Today 2012, 17, 774–783.
- Barbieri, L.; Bolognesi, A.; Valbonesi, P.; Polito, L.; Olivieri, F.; Stirpe, F. Polynucleotide: Adenosine glycosidase activity of immunotoxins containing ribosome-inactivating proteins. J. Drug Target. 2000, 8, 281–288.
- Qi, L.; Nett, T.M.; Allen, M.C.; Sha, X.; Harrison, G.S.; Frederick, B.A.; Crawford, E.D.; Glode, L.M. Binding and cytotoxicity of conjugated and recombinant fusion proteins targeted to the gonadotropin-releasing hormone receptor. Cancer Res. 2004, 64, 2090–2095.
- Frankel, A.; Welsh, P.; Richardson, J.; Robertus, J.D. Role of arginine 180 and glutamic acid 177 of ricin toxin A chain in enzymatic inactivation of ribosomes. Mol. Cell. Biol. 1990, 10, 6257–6263.
- Robertus, J.D.; Piatak, M.; Ferris, R.; Houston, L.L. Crystallization of ricin A chain obtained from a cloned gene expressed in Escherichia coli. J. Biol. Chem. 1987, 262, 19–20.
- Habuka, N.; Akiyama, K.; Tsuge, H.; Miyano, M.; Matsumoto, T.; Noma, M. Expression and secretion of Mirabilis antiviral protein in Escherichia coli and its inhibition of in vitro eukaryotic and prokaryotic protein synthesis. J. Biol. Chem. 1990, 265, 10988–10992.
- Kataoka, J.; Habuka, N.; Furuno, M.; Miyano, M.; Takanami, Y.; Koiwai, A. Expression of a pokeweed antiviral protein in Escherichia coli and its characterization. J. Biol. Chem. 1991, 266, 8426–8430.
- Legname, G.; Fossati, G.; Monzini, N.; Gromo, G.; Marcucci, F.; Mascagni, P.; Modena, D. Heterologous expression, purification, activity and conformational studies of different forms of dianthin 30. Biomed. Pept. Proteins Nucleic Acids 1995, 1, 61–68.
- Shaw, P.C.; Yun, M.H.; Zhu, R.H.; Ho, W.K.K.; Ng, T.B.; Yeung, H.W. Cloning of trichosanthin cDNA and its expression in Escherichia coli. Gene 1991, 97, 267–272.
- Bass, H.W.; Krawetz, J.E.; OBrian, G.R.; Zinselmeier, C.; Habben, J.E.; Boston, R.S. Maize ribosome-inactivating proteins (RIPs) with distinct expression patterns have similar requirements for proenzyme activation. J. Exp. Bot. 2004, 55, 2219–2233.
- Ding, G.B.; Wu, G.; Li, B.; Yang, P.; Li, Z. High-yield expression in Escherichia coli, biophysical characterization, and biological evaluation of plant toxin gelonin. 3 Biotech 2019, 9, 19.
- Hartley, M.R.; Legname, G.; Osborn, R.; Chen, Z.; Lord, J.M. Single-chain ribosome inactivating proteins from plants depurinate Escherichia coli 23S ribosomal RNA. FEBS Lett. 1991, 290, 65–68.
- Barthelemy, I.; Martineau, D.; Ong, M.; Matsunami, R.; Ling, N.; Benatti, L.; Cavallaro, U.; Soria, M.; Lappi, D.A. The expression of saporin, a ribosome-inactivating protein from the plant Saponaria officinalis, in Escherichia coli. J. Biol. Chem. 1993, 268, 6541–6548.
- Fabbrini, M.S.; Rappocciolo, E.; Carpani, D.; Solinas, M.; Valsasina, B.; Breme, U.; Cavallaro, U.; Nykjaer, A.; Rovida, E.; Legname, G.; et al. Characterization of a saporin isoform with lower ribosome-inhibiting activity. Biochem. J. 1997, 322, 719–727.
- Benatti, L.; Saccardo, M.B.; Dani, M.; Nitti, G.P.; Sassano, M.; Lorenzetti, R.; Lappi, D.A.; Soria, M. Nucleotide sequence of cDNA coding for saporin-6, a type-1 ribosome-inactivating protein from Saponaria officinalis. Eur. J. Biochem. 1989, 183, 465–470.
- Benatti, L.; Nitti, G.; Solinas, M.; Valsasina, B.; Vitale, A.; Ceriotti, A.; Soria, M.R. A Saporin-6 cDNA containing a precursor sequence coding for a carboxyl-terminal extension. FEBS Lett. 1991, 291, 285–288.
- Pittaluga, E.; Poma, A.; Tucci, A.; Spanò, L. Expression and characterisation in E. coli of mutant forms of saporin. J. Biotechnol. 2005, 117, 263–266.
- Günhan, E.; Swe, M.; Palazoglu, M.; Voss, J.C.; Chalupa, L.M. Expression and purification of cysteine introduced recombinant saporin. Protein Expr. Purif. 2008, 58, 203–209.
- Giansanti, F.; di Leandro, L.; Koutris, I.; Pitari, G.; Fabbrini, M.S.; Lombardi, A.; Flavel, D.J.; Flavell, S.U.; Gianni, S.; Ippoliti, R. Engineering a switchable toxin: The potential use of PDZ domains in the expression, targeting and activation of modified saporin variants. Protein Eng. Des. Sel. 2010, 23, 61–68.
- Giansanti, F.; Sabatini, D.; Pennacchio, M.R.; Scotti, S.; Angelucci, F.; Dhez, A.C.; Antonosante, A.; Cimini, A.; Giordano, A.; Ippoliti, R. PDZ Domain in the Engineering and Production of a Saporin Chimeric Toxin as a Tool for targeting Cancer Cells. J. Cell. Biochem. 2015, 116, 1256–1266.
- Weng, A.; Thakur, M.; von Mallinckrodt, B.; Beceren-Braun, F.; Gilabert-Oriol, R.; Wiesner, B.; Eichhorst, J.; Böttger, S.; Melzig, M.F.; Fuchs, H. Saponins modulate the intracellular trafficking of protein toxins. J. Control Release 2012, 164, 74–86.
- Zuppone, S.; Assalini, C.; Minici, C.; Botrugno, O.A.; Curnis, F.; Degano, M.; Corti, A.; Montorsi, F.; Salonia, A.; Vago, R.A. Novel RGD-4C-Saporin Conjugate Inhibits Tumor Growth in Mouse Models of Bladder Cancer. Front. Oncol. 2022, 12, 846958.
- Lombardi, A.; Bursomanno, S.; Lopardo, T.; Traini, R.; Colombatti, M.; Ippoliti, R.; Flavell, D.J.; Flavell, S.U.; Ceriotti, A.; Fabbrini, M.S. Pichia pastoris as a host for secretion of toxic saporin chimeras. FASEB J. 2010, 24, 253–265.
- Mattanovich, D.; Branduardi, P.; Dato, L.; Gasser, B.; Sauer, M.; Porro, D. Recombinant protein production in yeasts. In Methods in Molecular Biology (Clifton, N.J.); Humana Press: Totowa, NJ, USA, 2012; Volume 824, pp. 329–358.
- Heisler, I.; Keller, J.; Tauber, R.; Sutherland, M.; Fuchs, H. A cleavableadapter to reduce nonspecific cytotoxicity of recombinant immuno-toxins. Int. J. Cancer 2003, 103, 277–282.
- Vitetta, E.S. Immunotoxins and vascular leak syndrome. Cancer J. 2000, 6, 218–224.
- Chaudhary, V.K.; Gallo, M.G.; FitzGerald, D.J.; Pastan, I. A recombinant single-chain immunotoxin composed of anti-Tac variable regions and a truncated diphtheria toxin. Proc. Natl. Acad. Sci. USA 1990, 87, 9491–9494.
- Madhumathi, J.; Verma, R.S. Therapeutic targets and recent advances in protein immunotoxins. Curr. Opin. Microbiol. 2012, 15, 300–309.
- Fabbrini, M.S.; Flavell, D.J.; Ippoliti, R. Plant protein toxins: Structure, function and biotechnological applications. In Bacterial, Plant and Animal Toxins; Ascenzi, P.P.F., Visca, P., Eds.; Research Signpost: Kerala, India, 2003; pp. 66–69.
- Fabbrini, M.S.; Carpani, D.; Bello-Rivero, I.; Soria, M.R. The amino-terminal fragment of human urokinase directs a recombinant chimeric toxin to target cells: Internalization is toxin mediated. FASEB J. 1997, 11, 1169–1176.
- Della Cristina, P.; Castagna, M.; Lombardi, A.; Barison, E.; Tagliabue, G.; Ceriotti, A.; Koutris, I.; Di Leandro, L.; Giansanti, F.; Vago, R.; et al. Systematic comparison of single-chain Fv antibody-fusion toxin constructs containing Pseudomonas Exotoxin A or saporin produced in different microbial expression systems. Microb. Cell Fact. 2015, 14, 19.
- Errico Provenzano, A.; Posteri, R.; Giansanti, F.; Angelucci, F.; Flavell, S.U.; Flavell, D.J.; Fabbrini, M.S.; Porro, D.; Ippoliti, R.; Ceriotti, A.; et al. Optimization of construct design and fermentation strategy for the production of bioactive ATF-SAP, a saporin based anti-tumoral uPAR-targeted chimera. Microb. Cell Fact. 2016, 15, 194.
- Marshall, R.S.; D’Avila, F.; Di Cola, A.; Traini, R.; Spano, L.; Fabbrini, M.S.; Ceriotti, A. Signal peptide-regulated toxicity of a plant ribosome-inactivating protein during cell stress. Plant J. 2011, 65, 218–229.
- Frigerio, L.; Vitale, A.; Lord, J.M.; Ceriotti, A.; Roberts, L.M. Free ricin A chain, proricin, and native toxin have different cellular fates when expressed in tobacco protoplasts. J. Biol. Chem. 1998, 273, 14194–14199.
- Krishnan, R.; McDonald, K.A.; Dandekar, A.M.; Jackman, A.P.; Falk, B. Expression of recombinant trichosanthin, a ribosome-inactivating protein, in transgenic tobacco. J. Biotechnol. 2002, 97, 69–88.
- Zarovni, N.; Vago, R.; Soldà, T.; Monaco, L.; Fabbrini, M.S. Saporin as a novel suicide gene in anticancer gene therapy. Cancer Gene Ther. 2007, 14, 165–173.
- Zarovni, N.; Vago, R.; Fabbrini, M.S. Saporin suicide gene therapy. Methods Mol. Biol. 2009, 542, 261–283.
- Min, K.A.; He, H.; Yang, V.C.; Shin, M.C. Construction and characterization of gelonin and saporin plasmids for toxic gene-based cancer therapy. Arch. Pharm. Res. 2016, 39, 677–686.
- Salvioni, L.; Zuppone, S.; Andreata, F.; Monieri, M.; Mazzucchelli, S.; Di Carlo, C.; Morelli, L.; Cordiglieri, C.; Donnici, L.; De Francesco, R.; et al. Nanoparticle-Mediated Suicide Gene Therapy for Triple Negative Breast Cancer Treatment. Adv. Therap. 2020, 3, 2000007.
- di Leandro, L.; Giansanti, F.; Mei, S.; Ponziani, S.; Colasante, M.; Ardini, M.; Angelucci, F.; Pitari, G.; d’Angelo, M.; Cimini, A.; et al. Aptamer-Driven Toxin Gene Delivery in U87 Model Glioblastoma Cells. Front. Pharmacol. 2021, 12, 588306.
- Zhou, Y.; Li, X.-P.; Kahn, J.N.; Tumer, N.E. Functional Assays for Measuring the Catalytic Activity of Ribosome Inactivating Proteins. Toxins 2018, 10, 240.
- Shi, W.W.; Tang, Y.S.; Sze, S.Y.; Zhu, Z.N.; Wong, K.B.; Shaw, P.C. Crystal Structure of Ribosome-Inactivating Protein Ricin A Chain in Complex with the C-Terminal Peptide of the Ribosomal Stalk Protein P2. Toxins 2016, 8, 296.
- Watanabe, K.; Kawasaki, T.; Sako, N.; Funatsu, G. Actions of pokeweed antiviral protein on virus-infected protoplasts. Biosci. Biotechnol. Biochem. 1997, 61, 994–997.
- Gilabert-Oriol, R.; Weng, A.; Mallinckrodt, B.; Melzig, M.F.; Fuchs, H.; Thakur, M. Immunotoxins constructed with ribosome-inactivating proteins and their enhancers: A lethal cocktail with tumor specific efficacy. Curr. Pharm. Des. 2014, 20, 6584–6643.
- Bortolotti, M.; Polito, L.; Bolognesi, A. Toxin and Immunotoxin Based Therapeutic Approaches. Toxins 2022, 14, 63.
- Polito, L.; Djemil, A.; Bortolotti, M. Plant Toxin-Based Immunotoxins for Cancer Therapy: A Short Overview. Biomedicines 2016, 4, 12.
- Słomińska-Wojewódzka, M.; Sandvig, K. Ricin and Ricin-Containing Immunotoxins: Insights into Intracellular Transport and Mechanism of action in Vitro. Antibodies 2013, 2, 236–269.
- Schnell, R.; Borchmann, P.; Staak, J.O.; Schindler, J.; Ghetie, V.; Vitetta, E.S.; Engert, A. Clinical evaluation of ricin A-chain immunotoxins in patients with Hodgkin’s lymphoma. Ann. Oncol. 2003, 14, 729–736.
- Messmann, R.A.; Vitetta, E.S.; Headlee, D.; Senderowicz, A.M.; Figg, W.D.; Schindler, J.; Michiel, D.F.; Creekmore, S.; Steinberg, S.M.; Kohler, D.; et al. A phase I study of combination therapy with immunotoxins IgG-HD37-deglycosylated ricin A chain (dgA) and IgG-RFB4-dgA (Combotox) in patients with refractory CD19(+), CD22(+) B cell lymphoma. Clin. Cancer Res. 2000, 6, 1302–1313.
- LoRusso, P.M.; Lomen, P.L.; Redman, B.G.; Poplin, E.; Bander, J.J.; Valdivieso, M. Phase I study of monoclonal antibody-ricin A chain immunoconjugate Xomazyme-791 in patients with metastatic colon cancer. Am. J. Clin. Oncol. 1995, 18, 307–312.
- Engert, A.; Diehl, V.; Schnell, R.; Radszuhn, A.; Hatwig, M.T.; Drillich, S.; Schön, G.; Bohlen, H.; Tesch, H.; Hansmann, M.L.; et al. A phase-I study of an anti-CD25 ricin A-chain immunotoxin (RFT5-SMPT-dgA) in patients with refractory Hodgkin’s lymphoma. Blood 1997, 89, 403–410.
- de Virgilio, M.; Degryse, B. Harnessing the Destructive Power of Ricin to Fight Human Cancer. In Ricin Toxin; John, W.C., Ed.; Bentham Science Publishers: Soest, The Netherlands, 2014; pp. 208–237.
- Olsnes, S.; Pihl, A. Molecular Action of Toxins and Viruses; Cohen, P., Van Heyningen, S., Eds.; Elsevier Scientific Publishing Co., Inc.: New York, NY, USA, 1982; pp. 51–105.
- Naemi, A.A.; Salmanian, A.H.; Noormohammadi, Z.; Amani, J. A novel EGFR-specific recombinant ricin-panitumumab (scFv) immunotoxin against breast and colorectal cancer cell lines; in silico and in vitro analyses. Eur. J. Pharmacol. 2023, 955, 175894.
- Park, S.G.; Kim, H.; Jun, H.; Choi, S.Y.; Kim, E.; Kang, S. Directing ricin-based immunotoxins with targeting affibodies and KDEL signal peptide to cancer cells effectively induces apoptosis and tumor suppression. J. Nanobiotechnology 2022, 20, 387.
- Mlsna, D.; Monzingo, A.F.; Katzin, B.J.; Ernst, S.; Robertus, J.D. Structure of recombinant ricin A chain at 2.3 Å. Protein Sci. 1993, 2, 429–435.
- O’Hare, M.; Roberts, L.M.; Thorpe, P.E.; Watson, G.J.; Prior, B.; Lord, J.M. Expression of ricin a chain in Escherichia coli. FEBS Lett. 1987, 216, 73–78.
- FitzGerald, D.J.; Bjorn, M.J.; Ferris, R.J.; Winkelhake, J.L.; Frankel, A.E.; Hamilton, T.C.; Ozols, R.F.; Willingham, M.C.; Pastan, I. Antitumor activity of an immunotoxin in a nude mouse model of human ovarian cancer. Cancer Res. 1987, 47, 1407–1410.
- Li, B.Y.; Ramakrishnan, S. Recombinant hybrid toxin with dual enzymatic activities. Potential use in preparing highly effective immunotoxins. J. Biol. Chem. 1994, 269, 2652–2658.
- Zhan, J.; Ge, L.; Shen, J.; Wang, K.; Zheng, S. A trans-Golgi network retention signal YQRL fused to ricin A chain significantly enhances its cytotoxicity. Biochem. Biophys. Res. Commun. 2004, 313, 1053–1057.
- Zhan, J.; Stayton, P.; Press, O.W. Modification of ricin A chain, by addition of endoplasmic reticulum (KDEL) or Golgi (YQRL) retention sequences, enhances its cytotoxicity and translocation. Cancer Immunol. Immunother. 1998, 46, 55–60.
- Tagge, E.; Chandler, J.; Tang, B.L.; Hong, W.; Willingham, M.C.; Frankel, A. Cytotoxicity of KDEL-terminated ricin toxins correlates with distribution of the KDEL receptor in the Golgi. J. Histochem. Cytochem. 1996, 44, 159–165.
- Mahmoudi, R.; Dianat-Moghadam, H.; Poorebrahim, M.; Siapoush, S.; Poortahmasebi, V.; Salahlou, R.; Rahmati, M. Recombinant immunotoxins development for HER2-based targeted cancer therapies. Cancer Cell Int. 2021, 21, 470.
- Westby, M.; Argent, R.H.; Pitcher, C.; Lord, J.M.; Roberts, L.M. Preparation and characterization of recombinant proricin containing an alternative protease-sensitive linker sequence. Bioconjugate Chem. 1992, 3, 375–381.
- Kreitman, R.J.; Pastan, I. Recombinant toxins. Adv. Pharmacol. 1994, 28, 193–219.
- Benhar, I.; Pastan, I. Cloning, expression and characterization of the Fv fragments of the anti-carbohydrate mAbs B1 and B5 as single-chain immunotoxins. Protein Eng. 1994, 7, 1509–1515.
- Sørensen, H.P.; Mortensen, K.K. Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb. Cell Fact. 2005, 4, 1.
- Hassan, Y.; Ogg, S. Gene cloning and construction of prokaryotic and plant expression vectors of RICIN-A-Chain/PAP-S1 fusion protein and its inhibition of protein synthesis. bioRxiv 2016.
- Hassan, Y.; Ogg, S. Expression of Pokeweed Antiviral Protein Isoform S1 (PAP-S1) And of Ricin-A-Chain/PAP-S1 Novel Fusion Protein (RTA/PAP-S1) In Escherichia coli And Their Comparative Inhibition of Protein Synthesis In Vitro. bioRxiv 2017.
- Hassan, Y.; Ogg, S.; Ge, H. Expression of novel fusion antiviral proteins ricin a chain-pokeweed antiviral proteins (RTA-PAPs) in Escherichia coli and their inhibition of protein synthesis and of hepatitis B virus in vitro. BMC Biotechnol. 2018, 18, 47.
- Cook, J.P.; Savage, P.M.; Lord, J.M.; Roberts, L.M. Biologically active interleukin 2-ricin A chain fusion proteins may require intracellular proteolytic cleavage to exhibit a cytotoxic effect. Bioconjugate Chem. 1993, 4, 440–447.
- Asrorov, A.M.; Gu, Z.; Min, K.A.; Shin, M.C.; Huang, Y. Advances on Tumor-Targeting Delivery of Cytotoxic Proteins. ACS Pharmacol. Transl. Sci. 2019, 3, 107–118.
- Bortolotti, M.; Bolognesi, A.; Polito, L. Bouganin, an Attractive Weapon for Immunotoxins. Toxins 2018, 10, 323.
- Petrosini, L.; De Bartolo, P.; Cutuli, D. Neurotoxic Effects, Mechanisms, and Outcome of 192-IgG Saporin. In Handbook of Neurotoxicity; Kostrzewa, R., Ed.; Springer: New York, NY, USA, 2014.
- Bolognesi, A.; Polito, L.; Olivieri, F.; Valbonesi, P.; Barbieri, L.; Battelli, M.G.; Carusi, M.V.; Benvenuto, E.; Del Vecchio Blanco, F.; Di Maro, A.; et al. New Ribosome-Inactivating Proteins with Polynucleotide: Adenosine Glycosidase and Antiviral Activities from Basella Rubra, L. and Bougainvillea Spectabilis Willd. Planta 1997, 203, 422–429.
- Den Hartog, M.T.; Lubelli, C.; Boon, L.; Heerkens, S.; Ortiz Buijsse, A.P.; de Boer, M.; Stirpe, F. Cloning and Expression of cDNA Coding for Bouganin. Eur. J. Biochem. 2002, 269, 1772–1779.
- Fermani, S.; Tosi, G.; Farini, V.; Polito, L.; Falini, G.; Ripamonti, A.; Barbieri, L.; Chambery, A.; Bolognesi, A. Structure/Function Studies on Two Type 1 Ribosome Inactivating Proteins: Bouganin and Lychnin. J. Struct. Biol. 2009, 168, 278–287.
- Cizeau, J.; Grenkow, D.M.; Brown, J.G.; Entwistle, J.; MacDonald, G.C. Engineering and Biological Characterization of VB6-845, an Anti-EpCAM Immunotoxin Containing a T-Cell Epitope-Depleted Variant of the Plant Toxin Bouganin. J. Immunother. 2009, 32, 574–584.
- Dillon, R.L.; Chooniedass, S.; Premsukh, A.; Adams, G.P.; Entwistle, J.; MacDonald, G.C.; Cizeau, J. Trastuzumab-deBouganin Conjugate Overcomes Multiple Mechanisms of T-DM1 Drug Resistance. J. Immunother. 2016, 39, 117–126.
- Chooniedass, S.; Dillon, R.L.; Premsukh, A.; Adams, G.P.; MacDonald, G.C.; Cizeau, J. Abstract 79: Trastuzumab and C6.5 Diabody Armed with deBouganin Overcome Drug Resistance to ADCs Comprised of Anti-Microtubule Agents. Cancer Res. 2017, 77, 79.
- Kowalski, M.; Brazas, L.; Zaretsky, R.; Rasamoelisolo, M.; MacDonald, G.; Cuthbert, W.; Glover, N. A Phase I Study of VB6–845, an Anti-EpCAM Fusion Protein Targeting Advanced Solid Tumours of Epithelial Origin: Preliminary Results. J. Clin. Oncol. 2008, 26, 14663.
- Silva, A.L.; Goto, L.S.; Dinarte, A.R.; Hansen, D.; Moreira, R.A.; Beltramini, L.M.; Araújo, A.P. Pulchellin, a Highly Toxic Type 2 Ribosome-inactivating Protein from Abrus Pulchellus: Cloning, Heterologous Expression of A-chain and Structural Studies. FEBS J. 2005, 272, 1201–1210.
- Castilho, P.V.; Goto, L.S.; Roberts, L.M.; Araújo, A.P.U. Isolation and Characterization of Four Type 2 Ribosome Inactivating Pulchellin Isoforms from Abrus Pulchellus Seeds. FEBS J. 2008, 275, 948–959.
- Sdraeian, M.; Guimarães, F.E.G.; Araújo, A.P.U.; Worthylake, D.K.; LeCour, L.J.; Pincus, S.H. Selective Cytotoxicity of a Novel Immunotoxin Based on Pulchellin A Chain for Cells Expressing HIV Envelope. Sci. Rep. 2017, 7, 7579.
